A hydrogenotrophic Sulfurimonas is globally abundant in deep-sea oxygen-saturated hydrothermal plumes
- PMID: 36894632
- PMCID: PMC10066037
- DOI: 10.1038/s41564-023-01342-w
A hydrogenotrophic Sulfurimonas is globally abundant in deep-sea oxygen-saturated hydrothermal plumes
Abstract
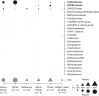
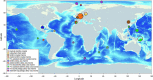
Members of the bacterial genus Sulfurimonas (phylum Campylobacterota) dominate microbial communities in marine redoxclines and are important for sulfur and nitrogen cycling. Here we used metagenomics and metabolic analyses to characterize a Sulfurimonas from the Gakkel Ridge in the Central Arctic Ocean and Southwest Indian Ridge, showing that this species is ubiquitous in non-buoyant hydrothermal plumes at Mid Ocean Ridges across the global ocean. One Sulfurimonas species, USulfurimonas pluma, was found to be globally abundant and active in cold (<0-4 °C), oxygen-saturated and hydrogen-rich hydrothermal plumes. Compared with other Sulfurimonas species, US. pluma has a reduced genome (>17%) and genomic signatures of an aerobic chemolithotrophic metabolism using hydrogen as an energy source, including acquisition of A2-type oxidase and loss of nitrate and nitrite reductases. The dominance and unique niche of US. pluma in hydrothermal plumes suggest an unappreciated biogeochemical role for Sulfurimonas in the deep ocean.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures









References
-
- Inagaki F, Takai K, Kobayashi H, Nealson KH, Horikoshi K. Sulfurimonas autotrophica gen. nov., sp. nov., a novel sulfur-oxidizing e-proteobacterium isolated from hydrothermal sediments in the Mid-Okinawa Trough. Int. J. Syst. Evol. Microbiol. 2003;53:1801–1805. doi: 10.1099/ijs.0.02682-0. - DOI - PubMed
-
- Timmer-Ten Hoor A. A new type of thiosulphate oxidizing, nitrate reducing microorganism: Thiomicrospira denitrificans sp. nov. Neth. J. Sea Res. 1975;9:344–350. doi: 10.1016/0077-7579(75)90008-3. - DOI
-
- Cai L, Shao M, Zhang T. Non-contiguous finished genome sequence and description of Sulfurimonas hongkongensis sp. nov., a strictly anaerobic denitrifying, hydrogen- and sulfur-oxidizing chemolithoautotroph isolated from marine sediment. Stand. Genom. Sci. 2014;9:1302–1310. doi: 10.4056/sigs.4948668. - DOI - PMC - PubMed
-
- Wang S, Jiang L, Liu X, Yang S, Shao Z. Sulfurimonas xiamenensis sp. nov. and Sulfurimonas lithotrophica sp. nov., hydrogen- and sulfur-oxidizing chemolithoautotrophs within the Epsilonproteobacteria isolated from coastal sediments, and an emended description of the genus Sulfurimonas. Int. J. Syst. Evol. Microbiol. 2020;70:2657–2663. doi: 10.1099/ijsem.0.004087. - DOI - PubMed
-
- Takai K, et al. Sulfurimonas paralvinellae sp. nov., a novel mesophilic, hydrogen- and sulfur-oxidizing chemolithoautotroph within the Epsilonproteobacteria isolated from a deep-sea hydrothermal vent polychaete nest, reclassification of Thiomicrospira denitrificans as Sulfurimonas denitrificans comb. nov. and emended description of the genus Sulfurimonas. Int. J. Syst. Evol. Microbiol. 2006;56:1725–1733. doi: 10.1099/ijs.0.64255-0. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

