YgfB increases β-lactam resistance in Pseudomonas aeruginosa by counteracting AlpA-mediated ampDh3 expression
- PMID: 36894667
- PMCID: PMC9998450
- DOI: 10.1038/s42003-023-04609-4
YgfB increases β-lactam resistance in Pseudomonas aeruginosa by counteracting AlpA-mediated ampDh3 expression
Abstract
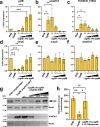
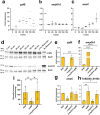
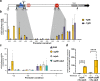
YgfB-mediated β-lactam resistance was recently identified in multi drug resistant Pseudomonas aeruginosa. We show that YgfB upregulates expression of the β-lactamase AmpC by repressing the function of the regulator of the programmed cell death pathway AlpA. In response to DNA damage, the antiterminator AlpA induces expression of the alpBCDE autolysis genes and of the peptidoglycan amidase AmpDh3. YgfB interacts with AlpA and represses the ampDh3 expression. Thus, YgfB indirectly prevents AmpDh3 from reducing the levels of cell wall-derived 1,6-anhydro-N-acetylmuramyl-peptides, required to induce the transcriptional activator AmpR in promoting the ampC expression and β-lactam resistance. Ciprofloxacin-mediated DNA damage induces AlpA-dependent production of AmpDh3 as previously shown, which should reduce β-lactam resistance. YgfB, however, counteracts the β-lactam enhancing activity of ciprofloxacin by repressing ampDh3 expression and lowering the benefits of this drug combination. Altogether, YgfB represents an additional player in the complex regulatory network of AmpC regulation.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interest.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

