Altered infective competence of the human gut microbiome in COVID-19
- PMID: 36894986
- PMCID: PMC9995755
- DOI: 10.1186/s40168-023-01472-7
Altered infective competence of the human gut microbiome in COVID-19
Abstract
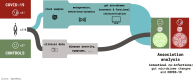
Background: Infections with SARS-CoV-2 have a pronounced impact on the gastrointestinal tract and its resident microbiome. Clear differences between severe cases of infection and healthy individuals have been reported, including the loss of commensal taxa. We aimed to understand if microbiome alterations including functional shifts are unique to severe cases or a common effect of COVID-19. We used high-resolution systematic multi-omic analyses to profile the gut microbiome in asymptomatic-to-moderate COVID-19 individuals compared to a control group.
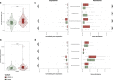
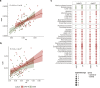
Results: We found a striking increase in the overall abundance and expression of both virulence factors and antimicrobial resistance genes in COVID-19. Importantly, these genes are encoded and expressed by commensal taxa from families such as Acidaminococcaceae and Erysipelatoclostridiaceae, which we found to be enriched in COVID-19-positive individuals. We also found an enrichment in the expression of a betaherpesvirus and rotavirus C genes in COVID-19-positive individuals compared to healthy controls.
Conclusions: Our analyses identified an altered and increased infective competence of the gut microbiome in COVID-19 patients. Video Abstract.
Keywords: COVID-19; Gut microbiome; Metagenomics; Metatranscriptomics; SARS-CoV-2.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- WHO Coronavirus (COVID-19) dashboard. https://covid19.who.int/. Accessed 10 Sept 2022.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

