Genome-wide analysis of R2R3-MYB transcription factors in Boehmeria nivea (L.) gaudich revealed potential cadmium tolerance and anthocyanin biosynthesis genes
- PMID: 36896232
- PMCID: PMC9989182
- DOI: 10.3389/fgene.2023.1080909
Genome-wide analysis of R2R3-MYB transcription factors in Boehmeria nivea (L.) gaudich revealed potential cadmium tolerance and anthocyanin biosynthesis genes
Abstract
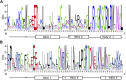
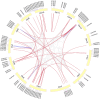
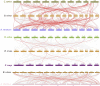
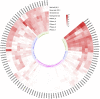
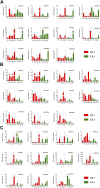
Gene family, especially MYB as one of the largest transcription factor family in plants, the study of its subfunctional characteristics is a key step in the study of plant gene function. The sequencing of ramie genome provides a good opportunity to study the organization and evolutionary characters of the ramie MYB gene at the whole genome level. In this study, a total of 105 BnGR2R3-MYB genes were identified from ramie genome and subsequently grouped into 35 subfamilies according to phylogeny divergence and sequences similarity. Chromosomal localization, gene structure, synteny analysis, gene duplication, promoter analysis, molecular characteristics and subcellular localization were accomplished using several bioinformatics tools. Collinearity analysis showed that the segmental and tandem duplication events is the dominant form of the gene family expansion, and duplications prominent in distal telomeric regions. Highest syntenic relationship was obtained between BnGR2R3-MYB genes and that of Apocynum venetum (88). Furthermore, transcriptomic data and phylogenetic analysis revealed that BnGMYB60, BnGMYB79/80 and BnGMYB70 might inhibit the biosynthesis of anthocyanins, and UPLC-QTOF-MS data further supported the results. qPCR and phylogenetic analysis revealed that the six genes (BnGMYB9, BnGMYB10, BnGMYB12, BnGMYB28, BnGMYB41, and BnGMYB78) were cadmium stress responsive genes. Especially, the expression of BnGMYB10/12/41 in roots, stems and leaves all increased more than 10-fold after cadmium stress, and in addition they may interact with key genes regulating flavonoid biosynthesis. Thus, a potential link between cadmium stress response and flavonoid synthesis was identified through protein interaction network analysis. The study thus provided significant information into MYB regulatory genes in ramie and may serve as a foundation for genetic enhancement and increased productivity.
Keywords: MYB TFs; anthocyanin biosynthesis; cadmium stress; expression profiles; protein interaction; ramie.
Copyright © 2023 Feng, Abubakar, Chen, Yu, Zhu, Chen, Gao, Wang, Mou and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Agarwal P., Mitra M., Banerjee S., Roy S. (2020). MYB4 transcription factor, a member of R2R3-subfamily of MYB domain protein, regulates cadmium tolerance via enhanced protection against oxidative damage and increases expression of PCS1 and MT1C in Arabidopsis. Plant Sci. 297, 110501. 10.1016/j.plantsci.2020.110501 - DOI - PubMed
LinkOut - more resources
Full Text Sources

