De novo Assembly and Comparative Analyses of Mitochondrial Genomes in Piperales
- PMID: 36896589
- PMCID: PMC10036691
- DOI: 10.1093/gbe/evad041
De novo Assembly and Comparative Analyses of Mitochondrial Genomes in Piperales
Abstract
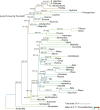
The mitochondrial genome of Liriodendron tulipifera exhibits many ancestral angiosperm features and a remarkably slow evolutionary rate, while mitochondrial genomes of other magnoliids remain yet to be characterized. We assembled nine new mitochondrial genomes, representing all genera of perianth-bearing Piperales, as well as for a member of the sister clade: three complete or nearly complete mitochondrial genomes from Aristolochiaceae and six additional draft assemblies including Thottea, Asaraceae, Lactoridaceae, and Hydnoraceae. For comparative purpose, a complete mitochondrial genome was assembled for Saururus, a member of the perianth-less Piperales. The average number of short repeats (50-99 bp) was much larger in genus Aristolochia than in other angiosperm mitochondrial genomes, and approximately 30% of repeats (<350 bp) were found to have the capacity to mediate recombination. We found mitochondrial genomes in perianth-bearing Piperales comprising conserved repertories of protein-coding genes and rRNAs but variable copy numbers of tRNA genes. We identified several shifts from cis- to trans-splicing of the Group II introns of nad1i728, cox2i373, and nad7i209. Two short regions of the cox1 and atp8 genes were likely derived from independent horizontal gene transfer events in perianth-bearing Piperales. We found biased enrichment of specific substitution types in different lineages of magnoliids and the Aristolochiaceae family showed the highest ratio of A:T > T:A substitutions of all other investigated angiosperm groups. Our study reports the first mitochondrial genomes for Piperales and uses this new information for a better understanding of the evolutionary patterns of magnoliids and angiosperms in general.
Keywords: Aristolochiaceae; HGT; Piperales; mitochondrial genome; mutation spectrum.
© The Author(s) 2023. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures




References
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources

