Fun30 nucleosome remodeller regulates white-to-opaque switching in Candida albicans
- PMID: 36896644
- PMCID: PMC10160231
- DOI: 10.3724/abbs.2023031
Fun30 nucleosome remodeller regulates white-to-opaque switching in Candida albicans
Abstract
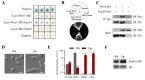
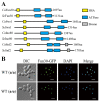
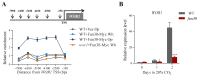
Candida albicans ( C. albicans) is an opportunistic pathogen in humans and possesses a white-opaque heritable switching system. Wor1 is a master regulator of white-opaque switching and is essential for opaque cell formation in C. albicans. However, the regulatory network of Wor1 in white-opaque switching is still vague. In this study, we obtain a series of Wor1-interacting proteins using LexA-Wor1 as bait. Among these proteins, function unknown now 30 (Fun30) interacts with Wor1 in vitro and in vivo. Fun30 expression is upregulated in opaque cells at the transcriptional and protein levels. Loss of FUN30 attenuates white-to-opaque switching, while ectopic expression of FUN30 significantly increases white-to-opaque switching in an ATPase activity-dependent manner. Furthermore, FUN30 upregulation is dependent on CO 2; loss of FLO8, a key CO 2-sensing transcriptional regulator, abolishes FUN30 upregulation. Interestingly, deletion of FUN30 affects the WOR1 expression regulation feedback loop. Thus, our results indicate that the chromatin remodeller Fun30 interacts with Wor1 and is required for WOR1 expression and opaque cell formation.
Keywords: Fun30; Wor1; chromatin remodeller; white-opaque switching.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Similar articles
-
The WOR1 5' untranslated region regulates white-opaque switching in Candida albicans by reducing translational efficiency.Mol Microbiol. 2015 Jul;97(1):125-38. doi: 10.1111/mmi.13014. Epub 2015 Apr 24. Mol Microbiol. 2015. PMID: 25831958 Free PMC article.
-
SUMOylation of Wor1 by a novel SUMO E3 ligase controls cell fate in Candida albicans.Mol Microbiol. 2015 Oct;98(1):69-89. doi: 10.1111/mmi.13108. Epub 2015 Jul 30. Mol Microbiol. 2015. PMID: 26112173
-
MTL-independent phenotypic switching in Candida tropicalis and a dual role for Wor1 in regulating switching and filamentation.PLoS Genet. 2013 Mar;9(3):e1003369. doi: 10.1371/journal.pgen.1003369. Epub 2013 Mar 21. PLoS Genet. 2013. PMID: 23555286 Free PMC article.
-
Regulation of white-opaque switching in Candida albicans.Med Microbiol Immunol. 2010 Aug;199(3):165-72. doi: 10.1007/s00430-010-0147-0. Med Microbiol Immunol. 2010. PMID: 20390300 Review.
-
White-opaque switching in Candida albicans: cell biology, regulation, and function.Microbiol Mol Biol Rev. 2024 Jun 27;88(2):e0004322. doi: 10.1128/mmbr.00043-22. Epub 2024 Mar 28. Microbiol Mol Biol Rev. 2024. PMID: 38546228 Free PMC article. Review.
References
-
- Kadosh D. Regulatory mechanisms controlling morphology and pathogenesis in Candida albicans . Curr Opin Microbiol. . 2019;52:27–34. doi: 10.1016/j.mib.2019.04.005. - DOI - PMC - PubMed
-
- Noble SM, Gianetti BA, Witchley JN. Candida albicans cell-type switching and functional plasticity in the mammalian host . Nat Rev Microbiol. . 2017;15:96–108. doi: 10.1038/nrmicro.2016.157. - DOI - PMC - PubMed
-
- Rikkerink EH, Magee BB, Magee PT. Opaque-white phenotype transition: a programmed morphological transition in Candida albicans . J Bacteriol. . 1988;170:895–899. doi: 10.1128/jb.170.2.895-899.1988. - DOI - PMC - PubMed
-
- Slutsky B, Staebell M, Anderson J, Risen L, Pfaller M, Soll DR. White-opaque transition: a second high-frequency switching system in Candida albicans . J Bacteriol. . 1987;169:189–197. doi: 10.1128/jb.169.1.189-197.1987. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

