StonPy: a tool to parse and query collections of SBGN maps in a graph database
- PMID: 36897014
- PMCID: PMC10017094
- DOI: 10.1093/bioinformatics/btad100
StonPy: a tool to parse and query collections of SBGN maps in a graph database
Abstract
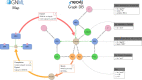
Summary: The systems biology graphical notation (SBGN) has become the de facto standard for the graphical representation of molecular maps. Having rapid and easy access to the content of large collections of maps is necessary to perform semantic or graph-based analysis of these resources. To this end, we propose StonPy, a new tool to store and query SBGN maps in a Neo4j graph database. StonPy notably includes a data model that takes into account all three SBGN languages and a completion module to automatically build valid SBGN maps from query results. StonPy is built as a library that can be integrated into other software and offers a command-line interface that allows users to easily perform all operations.
Availability and implementation: StonPy is implemented in Python 3 under a GPLv3 license. Its code and complete documentation are freely available from https://github.com/adrienrougny/stonpy.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2023. Published by Oxford University Press.
Figures