Palidis: fast discovery of novel insertion sequences
- PMID: 36897935
- PMCID: PMC10132078
- DOI: 10.1099/mgen.0.000917
Palidis: fast discovery of novel insertion sequences
Abstract
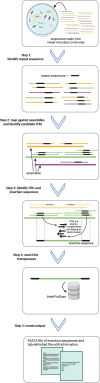
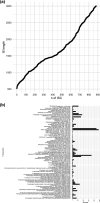
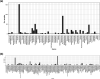
The diversity of microbial insertion sequences, crucial mobile genetic elements in generating diversity in microbial genomes, needs to be better represented in current microbial databases. Identification of these sequences in microbiome communities presents some significant problems that have led to their underrepresentation. Here, we present a bioinformatics pipeline called Palidis that recognizes insertion sequences in metagenomic sequence data rapidly by identifying inverted terminal repeat regions from mixed microbial community genomes. Applying Palidis to 264 human metagenomes identifies 879 unique insertion sequences, with 519 being novel and not previously characterized. Querying this catalogue against a large database of isolate genomes reveals evidence of horizontal gene transfer events across bacterial classes. We will continue to apply this tool more widely, building the Insertion Sequence Catalogue, a valuable resource for researchers wishing to query their microbial genomes for insertion sequences.
Keywords: antimicrobial resistance; horizontal gene transfer; insertion sequences; metagenome; mobile genetic element; transposon.
Conflict of interest statement
The authors declare no conflicting interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

