High-throughput genetic engineering of nonmodel and undomesticated bacteria via iterative site-specific genome integration
- PMID: 36897939
- PMCID: PMC10005180
- DOI: 10.1126/sciadv.ade1285
High-throughput genetic engineering of nonmodel and undomesticated bacteria via iterative site-specific genome integration
Abstract
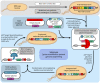
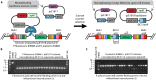
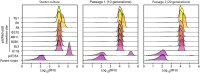
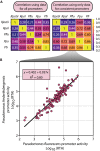
Efficient genome engineering is critical to understand and use microbial functions. Despite recent development of tools such as CRISPR-Cas gene editing, efficient integration of exogenous DNA with well-characterized functions remains limited to model bacteria. Here, we describe serine recombinase-assisted genome engineering, or SAGE, an easy-to-use, highly efficient, and extensible technology that enables selection marker-free, site-specific genome integration of up to 10 DNA constructs, often with efficiency on par with or superior to replicating plasmids. SAGE uses no replicating plasmids and thus lacks the host range limitations of other genome engineering technologies. We demonstrate the value of SAGE by characterizing genome integration efficiency in five bacteria that span multiple taxonomy groups and biotechnology applications and by identifying more than 95 heterologous promoters in each host with consistent transcription across environmental and genetic contexts. We anticipate that SAGE will rapidly expand the number of industrial and environmental bacteria compatible with high-throughput genetics and synthetic biology.
Figures


References
-
- V. M. Isabella, B. N. Ha, M. J. Castillo, D. J. Lubkowicz, S. E. Rowe, Y. A. Millet, C. L. Anderson, N. Li, A. B. Fisher, K. A. West, P. J. Reeder, M. M. Momin, C. G. Bergeron, S. E. Guilmain, P. F. Miller, C. B. Kurtz, D. Falb, Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria. Nat. Biotechnol. 36, 857–864 (2018). - PubMed
-
- V. Bahuguna, G. Bhatt, R. Maikhuri, D. Chandra, in Microbial Metatranscriptomics Belowground, M. Nath, D. Bhatt, P. Bhargava, D. K. Choudhary, Eds. (Springer Singapore, 2021), pp. 109–122.
-
- A. Z. Werner, R. Clare, T. D. Mand, I. Pardo, K. J. Ramirez, S. J. Haugen, F. Bratti, G. N. Dexter, J. R. Elmore, J. D. Huenemann, G. L. Peabody V, C. W. Johnson, N. A. Rorrer, D. Salvachúa, A. M. Guss, G. T. Beckham, Tandem chemical deconstruction and biological upcycling of poly(ethylene terephthalate) to β-ketoadipic acid by Pseudomonas putida KT2440. Metab. Eng. 67, 250–261 (2021). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

