Identification of Sodium- and Chloride-Sensitive Sites in the Slack Channel
- PMID: 36898835
- PMCID: PMC10089238
- DOI: 10.1523/JNEUROSCI.1365-22.2023
Identification of Sodium- and Chloride-Sensitive Sites in the Slack Channel
Abstract
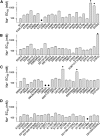
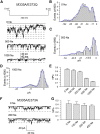
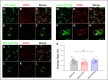
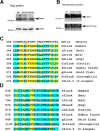
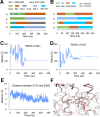
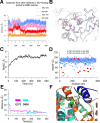
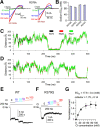
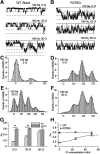
The Slack channel (KCNT1, Slo2.2) is a sodium-activated and chloride-activated potassium channel that regulates heart rate and maintains the normal excitability of the nervous system. Despite intense interest in the sodium gating mechanism, a comprehensive investigation to identify the sodium-sensitive and chloride-sensitive sites has been missing. In the present study, we identified two potential sodium-binding sites in the C-terminal domain of the rat Slack channel by conducting electrophysical recordings and systematic mutagenesis of cytosolic acidic residues in the rat Slack channel C terminus. In particular, by taking advantage of the M335A mutant, which results in the opening of the Slack channel in the absence of cytosolic sodium, we found that among the 92 screened negatively charged amino acids, E373 mutants could completely remove sodium sensitivity of the Slack channel. In contrast, several other mutants showed dramatic decreases in sodium sensitivity but did not abolish it altogether. Furthermore, molecular dynamics (MD) simulations performed at the hundreds of nanoseconds timescale revealed one or two sodium ions at the E373 position or an acidic pocket composed of several negatively charged residues. Moreover, the MD simulations predicted possible chloride interaction sites. By screening predicted positively charged residues, we identified R379 as a chloride interaction site. Thus, we conclude that the E373 site and the D863/E865 pocket are two potential sodium-sensitive sites, while R379 is a chloride interaction site in the Slack channel.SIGNIFICANCE STATEMENT The research presented here identified two distinct sodium and one chloride interaction sites located in the intracellular C-terminal domain of the Slack (Slo2.2, KCNT1) channel. Identification of the sites responsible for the sodium and chloride activation of the Slack channel sets its gating property apart from other potassium channels in the BK channel family. This finding sets the stage for future functional and pharmacological studies of this channel.
Keywords: KCNT1 channel; Slo2.2; chloride binding site; gating mechanism; sodium binding site.
Copyright © 2023 the authors.
Figures










References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
