Transcriptional Response to Hypoxia: The Role of HIF-1-Associated Co-Regulators
- PMID: 36899934
- PMCID: PMC10001186
- DOI: 10.3390/cells12050798
Transcriptional Response to Hypoxia: The Role of HIF-1-Associated Co-Regulators
Abstract
The Hypoxia Inducible Factor 1 (HIF-1) plays a major role in the cellular response to hypoxia by regulating the expression of many genes involved in adaptive processes that allow cell survival under low oxygen conditions. Adaptation to the hypoxic tumor micro-environment is also critical for cancer cell proliferation and therefore HIF-1 is also considered a valid therapeutical target. Despite the huge progress in understanding regulation of HIF-1 expression and activity by oxygen levels or oncogenic pathways, the way HIF-1 interacts with chromatin and the transcriptional machinery in order to activate its target genes is still a matter of intense investigation. Recent studies have identified several different HIF-1- and chromatin-associated co-regulators that play important roles in the general transcriptional activity of HIF-1, independent of its expression levels, as well as in the selection of binding sites, promoters and target genes, which, however, often depends on cellular context. We review here these co-regulators and examine their effect on the expression of a compilation of well-characterized HIF-1 direct target genes in order to assess the range of their involvement in the transcriptional response to hypoxia. Delineating the mode and the significance of the interaction between HIF-1 and its associated co-regulators may offer new attractive and specific targets for anticancer therapy.
Keywords: HIF-1; cancer; chromatin; hypoxia; transcriptional regulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
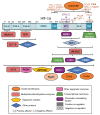

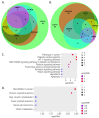
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

