Circular RNA Expression Signatures Provide Promising Diagnostic and Therapeutic Biomarkers for Chronic Lymphocytic Leukemia
- PMID: 36900344
- PMCID: PMC10000909
- DOI: 10.3390/cancers15051554
Circular RNA Expression Signatures Provide Promising Diagnostic and Therapeutic Biomarkers for Chronic Lymphocytic Leukemia
Abstract
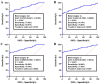
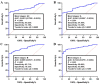
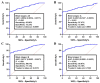
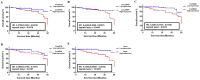
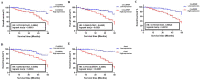
Chronic lymphocytic leukemia (CLL) is a known hematologic malignancy associated with a growing incidence and post-treatment relapse. Hence, finding a reliable diagnostic biomarker for CLL is crucial. Circular RNAs (circRNAs) represent a new class of RNA involved in many biological processes and diseases. This study aimed to define a circRNA-based panel for the early diagnosis of CLL. To this point, the list of the most deregulated circRNAs in CLL cell models was retrieved using bioinformatic algorithms and applied to the verified CLL patients' online datasets as the training cohort (n = 100). The diagnostic performance of potential biomarkers represented in individual and discriminating panels, was then analyzed between CLL Binet stages and validated in individual sample sets I (n = 220) and II (n = 251). We also estimated the 5-year overall survival (OS), introduced the cancer-related signaling pathways regulated by the announced circRNAs, and provided a list of possible therapeutic compounds to control the CLL. These findings show that the detected circRNA biomarkers exhibit better predictive performance compared to current validated clinical risk scales, and are applicable for the early detection and treatment of CLL.
Keywords: cancer; chronic lymphocytic leukemia; circular RNA; diagnosis; drug sensitivity prediction; prognosis.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- NIH Cancer Stat Facts: Leukemia—Chronic Lymphocytic Leukemia (CLL) [(accessed on 12 November 2022)]; Available online: https://seer.cancer.gov/statfacts/html/clyl.html.
-
- Swerdlow S.H., Campo E., Harris N.L., Jaffe E.S., Pileri S.A., Stein H., Thiele J., Vardiman J.W. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Volume 2 International Agency for Research on Cancer; Lyon, France: 2008.
-
- Bosch F., Montserrat E. Refining prognostic factors in chronic lymphocytic leukemia. Rev. Clin. Exp. Hematol. 2002;6:335–450. - PubMed
-
- Fais F., Ghiotto F., Hashimoto S., Sellars B., Valetto A., Allen S.L., Schulman P., Vinciguerra V.P., Rai K., Rassenti L.Z. Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J. Clin. Investig. 1998;102:1515–1525. doi: 10.1172/JCI3009. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

