Independent Validation of a Deep Learning nnU-Net Tool for Neuroblastoma Detection and Segmentation in MR Images
- PMID: 36900410
- PMCID: PMC10000775
- DOI: 10.3390/cancers15051622
Independent Validation of a Deep Learning nnU-Net Tool for Neuroblastoma Detection and Segmentation in MR Images
Abstract
Objectives: To externally validate and assess the accuracy of a previously trained fully automatic nnU-Net CNN algorithm to identify and segment primary neuroblastoma tumors in MR images in a large children cohort.
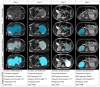
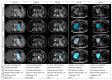
Methods: An international multicenter, multivendor imaging repository of patients with neuroblastic tumors was used to validate the performance of a trained Machine Learning (ML) tool to identify and delineate primary neuroblastoma tumors. The dataset was heterogeneous and completely independent from the one used to train and tune the model, consisting of 300 children with neuroblastic tumors having 535 MR T2-weighted sequences (486 sequences at diagnosis and 49 after finalization of the first phase of chemotherapy). The automatic segmentation algorithm was based on a nnU-Net architecture developed within the PRIMAGE project. For comparison, the segmentation masks were manually edited by an expert radiologist, and the time for the manual editing was recorded. Different overlaps and spatial metrics were calculated to compare both masks.
Results: The median Dice Similarity Coefficient (DSC) was high 0.997; 0.944-1.000 (median; Q1-Q3). In 18 MR sequences (6%), the net was not able neither to identify nor segment the tumor. No differences were found regarding the MR magnetic field, type of T2 sequence, or tumor location. No significant differences in the performance of the net were found in patients with an MR performed after chemotherapy. The time for visual inspection of the generated masks was 7.9 ± 7.5 (mean ± Standard Deviation (SD)) seconds. Those cases where manual editing was needed (136 masks) required 124 ± 120 s.
Conclusions: The automatic CNN was able to locate and segment the primary tumor on the T2-weighted images in 94% of cases. There was an extremely high agreement between the automatic tool and the manually edited masks. This is the first study to validate an automatic segmentation model for neuroblastic tumor identification and segmentation with body MR images. The semi-automatic approach with minor manual editing of the deep learning segmentation increases the radiologist's confidence in the solution with a minor workload for the radiologist.
Keywords: automatic segmentation; deep learning; external validation; independent validation; neuroblastic tumors; tumor segmentation.
Conflict of interest statement
The authors of this manuscript declare relationships with the following companies: QUIBIM SL.
Figures

References
-
- Desouza N.M., van der Lugt A., Deroose C.M., Alberich-Bayarri A., Bidaut L., Fournier L., Costaridou L., Oprea-Lager D.E., Kotter E., Smits M., et al. Standardised lesion segmentation for imaging biomarker quantitation: A consensus recommendation from ESR and EORTC. Insights Into Imaging. 2022;13:159. doi: 10.1186/s13244-022-01287-4. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

