Multifaceted Transcriptional Network of Estrogen-Related Receptor Alpha in Health and Disease
- PMID: 36901694
- PMCID: PMC10002233
- DOI: 10.3390/ijms24054265
Multifaceted Transcriptional Network of Estrogen-Related Receptor Alpha in Health and Disease
Abstract
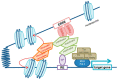
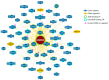
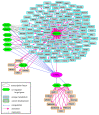
Estrogen-related receptors (ERRα, β and γ in mammals) are orphan members of the nuclear receptor superfamily acting as transcription factors. ERRs are expressed in several cell types and they display various functions in normal and pathological contexts. Amongst others, they are notably involved in bone homeostasis, energy metabolism and cancer progression. In contrast to other nuclear receptors, the activities of the ERRs are apparently not controlled by a natural ligand but they rely on other means such as the availability of transcriptional co-regulators. Here we focus on ERRα and review the variety of co-regulators that have been identified by various means for this receptor and their reported target genes. ERRα cooperates with distinct co-regulators to control the expression of distinct sets of target genes. This exemplifies the combinatorial specificity of transcriptional regulation that induces discrete cellular phenotypes depending on the selected coregulator. We finally propose an integrated view of the ERRα transcriptional network.
Keywords: cancer; metabolism; nuclear receptor; regulatory network; target gene; transcriptional regulator.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

