Back to Basics: A Simplified Improvement to Multiple Displacement Amplification for Microbial Single-Cell Genomics
- PMID: 36901710
- PMCID: PMC10002425
- DOI: 10.3390/ijms24054270
Back to Basics: A Simplified Improvement to Multiple Displacement Amplification for Microbial Single-Cell Genomics
Abstract
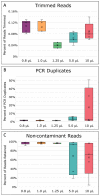
Microbial single-cell genomics (SCG) provides access to the genomes of rare and uncultured microorganisms and is a complementary method to metagenomics. Due to the femtogram-levels of DNA in a single microbial cell, sequencing the genome requires whole genome amplification (WGA) as a preliminary step. However, the most common WGA method, multiple displacement amplification (MDA), is known to be costly and biased against specific genomic regions, preventing high-throughput applications and resulting in uneven genome coverage. Thus, obtaining high-quality genomes from many taxa, especially minority members of microbial communities, becomes difficult. Here, we present a volume reduction approach that significantly reduces costs while improving genome coverage and uniformity of DNA amplification products in standard 384-well plates. Our results demonstrate that further volume reduction in specialized and complex setups (e.g., microfluidic chips) is likely unnecessary to obtain higher-quality microbial genomes. This volume reduction method makes SCG more feasible for future studies, thus helping to broaden our knowledge on the diversity and function of understudied and uncharacterized microorganisms in the environment.
Keywords: cell sorting; contact-free liquid dispenser; microbial dark matter; miniaturization; whole genome amplification.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- McDonald D., Price M.N., Goodrich J., Nawrocki E.P., Desantis T.Z., Probst A., Andersen G.L., Knight R., Hugenholtz P. An Improved Greengenes Taxonomy with Explicit Ranks for Ecological and Evolutionary Analyses of Bacteria and Archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

