Whole-Exome Sequencing and cfDNA Analysis Uncover Genetic Determinants of Melanoma Therapy Response in a Real-World Setting
- PMID: 36901733
- PMCID: PMC10002464
- DOI: 10.3390/ijms24054302
Whole-Exome Sequencing and cfDNA Analysis Uncover Genetic Determinants of Melanoma Therapy Response in a Real-World Setting
Abstract
Although several studies have explored the molecular landscape of metastatic melanoma, the genetic determinants of therapy resistance are still largely unknown. Here, we aimed to determine the contribution of whole-exome sequencing and circulating free DNA (cfDNA) analysis in predicting response to therapy in a consecutive real-world cohort of 36 patients, undergoing fresh tissue biopsy and followed during treatment. Although the underpowered sample size limited statistical analysis, samples from non-responders had higher copy number variations and mutations in melanoma driver genes compared to responders in the BRAF V600+ subset. In the BRAF V600- subset, Tumor Mutational Burden (TMB) was twice that in responders vs. non-responders. Genomic layout revealed commonly known and novel potential intrinsic/acquired resistance driver gene variants. Among these, RAC1, FBXW7, GNAQ mutations, and BRAF/PTEN amplification/deletion were present in 42% and 67% of patients, respectively. Both Loss of Heterozygosity (LOH) load and tumor ploidy were inversely associated with TMB. In immunotherapy-treated patients, samples from responders showed higher TMB and lower LOH and were more frequently diploid compared to non-responders. Secondary germline testing and cfDNA analysis proved their efficacy in finding germline predisposing variants carriers (8.3%) and following dynamic changes during treatment as a surrogate of tissue biopsy, respectively.
Keywords: BRAF V600; circulating free DNA; germline pathogenic variants; immunotherapy; loss of heterozygosity; melanoma; targeted therapy; tumor mutational burden; tumor ploidy; whole-exome sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
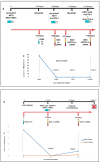

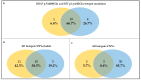
References
-
- Korn E.L., Liu P.-Y., Lee S.J., Chapman J.-A.W., Niedzwiecki D., Suman V.J., Moon J., Sondak V.K., Atkins M.B., Eisenhauer E.A., et al. Meta-Analysis of Phase II Cooperative Group Trials in Metastatic Stage IV Melanoma to Determine Progression-Free and Overall Survival Benchmarks for Future Phase II Trials. JCO. 2008;26:527–534. doi: 10.1200/JCO.2007.12.7837. - DOI - PubMed
MeSH terms
Substances
Grants and funding
- RF-2016-02362288/Ministero della Salute
- 5x1000 funds 2019-2020 to IRCCS Ospedale Policlinico San Martino/Ministero della Salute
- Ricerca Corrente 2020-2021 to IRCCS Ospedale Policlinico San Martino/Ministero della Salute
- Alliance Against Cancer to IRCCS Ospedale Policlinico San Martino/Alleanza Contro il Cancro
LinkOut - more resources
Full Text Sources
Medical
Research Materials

