Structural Basis of the Binding Mode of the Antineoplastic Compound Motixafortide (BL-8040) in the CXCR4 Chemokine Receptor
- PMID: 36901829
- PMCID: PMC10001991
- DOI: 10.3390/ijms24054393
Structural Basis of the Binding Mode of the Antineoplastic Compound Motixafortide (BL-8040) in the CXCR4 Chemokine Receptor
Abstract
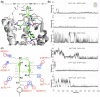
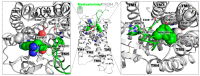
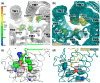
Modulation of the CXCL12-CXCR4 signaling axis is of the utmost importance due to its central involvement in several pathological disorders, including inflammatory diseases and cancer. Among the different currently available drugs that inhibit CXCR4 activation, motixafortide-a best-in-class antagonist of this GPCR receptor-has exhibited promising results in preclinical studies of pancreatic, breast, and lung cancers. However, detailed information on the interaction mechanism of motixafortide is still lacking. Here, we characterize the motixafortide/CXCR4 and CXCL12/CXCR4 protein complexes by using computational techniques including unbiased all-atom molecular dynamics simulations. Our microsecond-long simulations of the protein systems indicate that the agonist triggers changes associated with active-like GPCR conformations, while the antagonist favors inactive conformations of CXCR4. Detailed ligand-protein analysis indicates the importance of motixafortide's six cationic residues, all of which established charge-charge interactions with acidic CXCR4 residues. Furthermore, two synthetic bulky chemical moieties of motixafortide work in tandem to restrict the conformations of important residues associated with CXCR4 activation. Our results not only elucidate the molecular mechanism by which motixafortide interacts with the CXCR4 receptor and stabilizes its inactive states, but also provide essential information to rationally design CXCR4 inhibitors that preserve the outstanding pharmacological features of motixafortide.
Keywords: CXCR4; MD simulations; antineoplastic drugs; cancer; computational methods; motixafortide.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Bockorny B., Semenisty V., Macarulla T., Borazanci E., Wolpin B.M., Stemmer S.M., Golan T., Geva R., Borad M.J., Pedersen K.S., et al. BL-8040, a CXCR4 Antagonist, in Combination with Pembrolizumab and Chemotherapy for Pancreatic Cancer: The COMBAT Trial. Nat. Med. 2020;26:878–885. doi: 10.1038/s41591-020-0880-x. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

