Lessons Learnt from COVID-19: Computational Strategies for Facing Present and Future Pandemics
- PMID: 36901832
- PMCID: PMC10003049
- DOI: 10.3390/ijms24054401
Lessons Learnt from COVID-19: Computational Strategies for Facing Present and Future Pandemics
Abstract
Since its outbreak in December 2019, the COVID-19 pandemic has caused the death of more than 6.5 million people around the world. The high transmissibility of its causative agent, the SARS-CoV-2 virus, coupled with its potentially lethal outcome, provoked a profound global economic and social crisis. The urgency of finding suitable pharmacological tools to tame the pandemic shed light on the ever-increasing importance of computer simulations in rationalizing and speeding up the design of new drugs, further stressing the need for developing quick and reliable methods to identify novel active molecules and characterize their mechanism of action. In the present work, we aim at providing the reader with a general overview of the COVID-19 pandemic, discussing the hallmarks in its management, from the initial attempts at drug repurposing to the commercialization of Paxlovid, the first orally available COVID-19 drug. Furthermore, we analyze and discuss the role of computer-aided drug discovery (CADD) techniques, especially those that fall in the structure-based drug design (SBDD) category, in facing present and future pandemics, by showcasing several successful examples of drug discovery campaigns where commonly used methods such as docking and molecular dynamics have been employed in the rational design of effective therapeutic entities against COVID-19.
Keywords: CADD; COVID-19; SARS-CoV-2; SBDD; docking; homology modeling; molecular dynamics; pharmacophore; protein–ligand interaction fingerprints; rational drug design.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



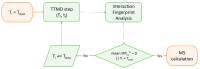
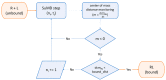
References
-
- Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., et al. The Species Severe Acute Respiratory Syndrome-Related Coronavirus: Classifying 2019-NCoV and Naming It SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. - DOI - PMC - PubMed
-
- WHO Director-General’s Remarks at the Media Briefing on 2019-NCoV on 11 February 2020. [(accessed on 5 December 2022)]. Available online: https://www.who.int/director-general/speeches/detail/who-director-genera....
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

