Protein Abundance of Drug Metabolizing Enzymes in Human Hepatitis C Livers
- PMID: 36901973
- PMCID: PMC10002520
- DOI: 10.3390/ijms24054543
Protein Abundance of Drug Metabolizing Enzymes in Human Hepatitis C Livers
Abstract
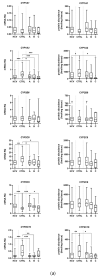
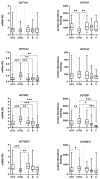
Hepatic drug metabolizing enzymes (DMEs), whose activity may be affected by liver diseases, are major determinants of drug pharmacokinetics. Hepatitis C liver samples in different functional states, i.e., the Child-Pugh class A (n = 30), B (n = 21) and C (n = 7) were analyzed for protein abundances (LC-MS/MS) and mRNA levels (qRT-PCR) of 9 CYPs and 4 UGTs enzymes. The protein levels of CYP1A1, CYP2B6, CYP2C8, CYP2C9, and CYP2D6 were not affected by the disease. In the Child-Pugh class A livers, a significant up-regulation of UGT1A1 (to 163% of the controls) was observed. The Child-Pugh class B was associated with down-regulation of the protein abundance of CYP2C19 (to 38% of the controls), CYP2E1 (to 54%), CYP3A4 (to 33%), UGT1A3 (to 69%), and UGT2B7 (to 56%). In the Child-Pugh class C livers, CYP1A2 was found to be reduced (to 52%). A significant trend in down-regulation of the protein abundance was documented for CYP1A2, CYP2C9, CYP3A4, CYP2E1, UGT2B7, and UGT2B15. The results of the study demonstrate that DMEs protein abundances in the liver are affected by hepatitis C virus infection and depend on the severity of the disease.
Keywords: drug metabolizing enzymes; hepatitis C; liver.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Drozdzik M., Busch D., Lapczuk J., Müller J., Ostrowski M., Kurzawski M., Oswald S. Protein Abundance of Clinically Relevant Drug-Metabolizing Enzymes in the Human Liver and Intestine: A Comparative Analysis in Paired Tissue Specimens. Clin. Pharmacol. Ther. 2018;104:515–524. doi: 10.1002/cpt.967. - DOI - PubMed
-
- European Medicines Agency 2005 Guideline on the Evaluation of the Pharmacokinetics of Medicinal Products in Patients with Impaired Hepatic Function. February 2005 [Online] [(accessed on 4 December 2022)]. Available online: https://www.ema.europa.eu/documents/scientific-guideline/guideline-evalu....
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

