Separase and Roads to Disengage Sister Chromatids during Anaphase
- PMID: 36902034
- PMCID: PMC10003635
- DOI: 10.3390/ijms24054604
Separase and Roads to Disengage Sister Chromatids during Anaphase
Abstract
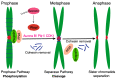
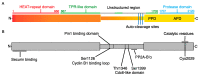
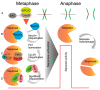
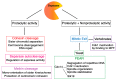
Receiving complete and undamaged genetic information is vital for the survival of daughter cells after chromosome segregation. The most critical steps in this process are accurate DNA replication during S phase and a faithful chromosome segregation during anaphase. Any errors in DNA replication or chromosome segregation have dire consequences, since cells arising after division might have either changed or incomplete genetic information. Accurate chromosome segregation during anaphase requires a protein complex called cohesin, which holds together sister chromatids. This complex unifies sister chromatids from their synthesis during S phase, until separation in anaphase. Upon entry into mitosis, the spindle apparatus is assembled, which eventually engages kinetochores of all chromosomes. Additionally, when kinetochores of sister chromatids assume amphitelic attachment to the spindle microtubules, cells are finally ready for the separation of sister chromatids. This is achieved by the enzymatic cleavage of cohesin subunits Scc1 or Rec8 by an enzyme called Separase. After cohesin cleavage, sister chromatids remain attached to the spindle apparatus and their poleward movement on the spindle is initiated. The removal of cohesion between sister chromatids is an irreversible step and therefore it must be synchronized with assembly of the spindle apparatus, since precocious separation of sister chromatids might lead into aneuploidy and tumorigenesis. In this review, we focus on recent discoveries concerning the regulation of Separase activity during the cell cycle.
Keywords: CDK1; Cyclin B1; Mad2; Sgo2; aneuploidy; chromosome division; cohesin; securin; segregation errors; separase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Splitting the chromosome: cutting the ties that bind sister chromatids.Science. 2000 May 26;288(5470):1379-85. doi: 10.1126/science.288.5470.1379. Science. 2000. PMID: 10827941 Review.
-
Splitting the nucleus: what's wrong with the tripartite ring model?Cold Spring Harb Symp Quant Biol. 2010;75:375-88. doi: 10.1101/sqb.2010.75.019. Epub 2011 Jan 5. Cold Spring Harb Symp Quant Biol. 2010. PMID: 21209385
-
Securin acetylation prevents precocious separase activation and premature sister chromatid separation.Curr Biol. 2024 Mar 25;34(6):1295-1308.e5. doi: 10.1016/j.cub.2024.02.038. Epub 2024 Mar 6. Curr Biol. 2024. PMID: 38452759
-
Cyclin A2 is required for sister chromatid segregation, but not separase control, in mouse oocyte meiosis.Cell Rep. 2012 Nov 29;2(5):1077-87. doi: 10.1016/j.celrep.2012.10.002. Epub 2012 Nov 1. Cell Rep. 2012. PMID: 23122964
-
[Activity of separase and its regulation].Yi Chuan. 2004 May;26(3):383-6. Yi Chuan. 2004. PMID: 15640025 Review. Chinese.
Cited by
-
The upregulation and transcriptional regulatory mechanisms of Extra spindle pole bodies like 1 in bladder cancer: An immunohistochemistry and high-throughput screening Evaluation.Heliyon. 2024 May 15;10(10):e31192. doi: 10.1016/j.heliyon.2024.e31192. eCollection 2024 May 30. Heliyon. 2024. PMID: 38813236 Free PMC article.
-
Eliminating separase inhibition reveals absence of robust cohesin protection in oocyte metaphase II.EMBO J. 2025 Aug 5. doi: 10.1038/s44318-025-00522-0. Online ahead of print. EMBO J. 2025. PMID: 40764838
-
Role of chromosomal cohesion and separation in aneuploidy and tumorigenesis.Cell Mol Life Sci. 2024 Feb 22;81(1):100. doi: 10.1007/s00018-024-05122-5. Cell Mol Life Sci. 2024. PMID: 38388697 Free PMC article. Review.
-
Live Cell Monitoring of Separase Activity, a Key Enzymatic Reaction for Chromosome Segregation, with Chimeric FRET-Based Molecular Sensor upon Cell Cycle Progression.Biosensors (Basel). 2024 Apr 15;14(4):192. doi: 10.3390/bios14040192. Biosensors (Basel). 2024. PMID: 38667185 Free PMC article.
-
Emerging roles of cohesin-STAG2 in cancer.Oncogene. 2025 Feb;44(5):277-287. doi: 10.1038/s41388-024-03221-y. Epub 2024 Nov 29. Oncogene. 2025. PMID: 39613934 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

