Transcription Factor ZmNAC20 Improves Drought Resistance by Promoting Stomatal Closure and Activating Expression of Stress-Responsive Genes in Maize
- PMID: 36902144
- PMCID: PMC10003513
- DOI: 10.3390/ijms24054712
Transcription Factor ZmNAC20 Improves Drought Resistance by Promoting Stomatal Closure and Activating Expression of Stress-Responsive Genes in Maize
Abstract
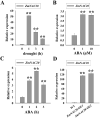
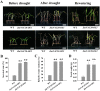
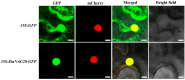
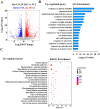
Drought is a major environmental threat that limits crop growth, development, and productivity worldwide. Improving drought resistance with genetic engineering methods is necessary to tackle global climate change. It is well known that NAC (NAM, ATAF and CUC) transcription factors play a critical role in coping with drought stress in plants. In this study, we identified an NAC transcription factor ZmNAC20, which regulates drought stress response in maize. ZmNAC20 expression was rapidly upregulated by drought and abscisic acid (ABA). Under drought conditions, the ZmNAC20-overexpressing plants had higher relative water content and survival rate than the wild-type maize inbred B104, suggesting that overexpression of ZmNAC20 improved drought resistance in maize. The detached leaves of ZmNAC20-overexpressing plants lost less water than those of wild-type B104 after dehydration. Overexpression of ZmNAC20 promoted stomatal closure in response to ABA. ZmNAC20 was localized in the nucleus and regulated the expression of many genes involved in drought stress response using RNA-Seq analysis. The study indicated that ZmNAC20 improved drought resistance by promoting stomatal closure and activating the expression of stress-responsible genes in maize. Our findings provide a valuable gene and new clues on improving crop drought resistance.
Keywords: ABA; ZmNAC20; drought resistance; stomatal closure; transcription factor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Genetic, molecular and physiological crosstalk during drought tolerance in maize (Zea mays): pathways to resilient agriculture.Planta. 2024 Aug 28;260(4):81. doi: 10.1007/s00425-024-04517-9. Planta. 2024. PMID: 39196449 Review.
-
Maize WRKY Transcription Factor ZmWRKY79 Positively Regulates Drought Tolerance through Elevating ABA Biosynthesis.Int J Mol Sci. 2021 Sep 18;22(18):10080. doi: 10.3390/ijms221810080. Int J Mol Sci. 2021. PMID: 34576244 Free PMC article.
-
ZmHB53, a Maize Homeodomain-Leucine Zipper I Transcription Factor Family Gene, Contributes to Abscisic Acid Sensitivity and Confers Seedling Drought Tolerance by Promoting the Activity of ZmPYL4.Plant Cell Environ. 2025 Jun;48(6):3829-3843. doi: 10.1111/pce.15394. Epub 2025 Jan 20. Plant Cell Environ. 2025. PMID: 39829370
-
Maize GOLDEN2-LIKE proteins enhance drought tolerance in rice by promoting stomatal closure.Plant Physiol. 2024 Jan 31;194(2):774-786. doi: 10.1093/plphys/kiad561. Plant Physiol. 2024. PMID: 37850886 Free PMC article.
-
Regulatory Gene Networks in Drought Stress Responses and Resistance in Plants.Adv Exp Med Biol. 2018;1081:189-214. doi: 10.1007/978-981-13-1244-1_11. Adv Exp Med Biol. 2018. PMID: 30288711 Review.
Cited by
-
Genetic, molecular and physiological crosstalk during drought tolerance in maize (Zea mays): pathways to resilient agriculture.Planta. 2024 Aug 28;260(4):81. doi: 10.1007/s00425-024-04517-9. Planta. 2024. PMID: 39196449 Review.
-
Identification of ZmSNAC06, a Maize NAC Family Transcription Factor with Multiple Transcripts Conferring Drought Tolerance in Arabidopsis.Plants (Basel). 2024 Dec 24;14(1):12. doi: 10.3390/plants14010012. Plants (Basel). 2024. PMID: 39795271 Free PMC article.
-
The overexpression of ascorbate peroxidase 2 (APX2) gene improves drought tolerance in maize.Mol Breed. 2025 Feb 15;45(2):27. doi: 10.1007/s11032-025-01548-2. eCollection 2025 Feb. Mol Breed. 2025. PMID: 39963376
-
Transcriptome and Metabolome Analyses Reveal the Molecular Mechanisms of Albizia odoratissima's Response to Drought Stress.Plants (Basel). 2024 Sep 29;13(19):2732. doi: 10.3390/plants13192732. Plants (Basel). 2024. PMID: 39409602 Free PMC article.
-
The Functions of an NAC Transcription Factor, GhNAC2-A06, in Cotton Response to Drought Stress.Plants (Basel). 2023 Nov 2;12(21):3755. doi: 10.3390/plants12213755. Plants (Basel). 2023. PMID: 37960109 Free PMC article.
References
MeSH terms
Substances
Grants and funding
- 222102110070/Key Project and Special Foundation of Research, Development, and Promotion in Henan Province
- 31800165/National Natural Science Foundation of China
- 32000147/National Natural Science Foundation of China
- 30500686/Research start-up fund to young talents of Henan Agricultural University
- 221RTSTHN0230/Program for Innovative Research Team (in Science and Technology) in University of Henan Province
LinkOut - more resources
Full Text Sources
Other Literature Sources

