When Just One Phosphate Is One Too Many: The Multifaceted Interplay between Myc and Kinases
- PMID: 36902175
- PMCID: PMC10003727
- DOI: 10.3390/ijms24054746
When Just One Phosphate Is One Too Many: The Multifaceted Interplay between Myc and Kinases
Abstract
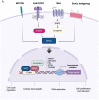
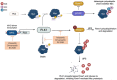
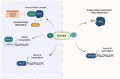
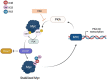
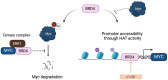
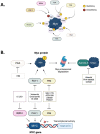
Myc transcription factors are key regulators of many cellular processes, with Myc target genes crucially implicated in the management of cell proliferation and stem pluripotency, energy metabolism, protein synthesis, angiogenesis, DNA damage response, and apoptosis. Given the wide involvement of Myc in cellular dynamics, it is not surprising that its overexpression is frequently associated with cancer. Noteworthy, in cancer cells where high Myc levels are maintained, the overexpression of Myc-associated kinases is often observed and required to foster tumour cells' proliferation. A mutual interplay exists between Myc and kinases: the latter, which are Myc transcriptional targets, phosphorylate Myc, allowing its transcriptional activity, highlighting a clear regulatory loop. At the protein level, Myc activity and turnover is also tightly regulated by kinases, with a finely tuned balance between translation and rapid protein degradation. In this perspective, we focus on the cross-regulation of Myc and its associated protein kinases underlying similar and redundant mechanisms of regulation at different levels, from transcriptional to post-translational events. Furthermore, a review of the indirect effects of known kinase inhibitors on Myc provides an opportunity to identify alternative and combined therapeutic approaches for cancer treatment.
Keywords: Aurora-A; Aurora-B; BRD4; GSK-3; Myc; PIM; PKA; PLK1; kinase inhibitors; kinases.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Schwab M., Alitalo K., Klempnauer K.-H., Varmus H.E., Bishop J.M., Gilbert F., Brodeur G., Goldstein M., Trent J. Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature. 1983;305:245–248. doi: 10.1038/305245a0. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

