The Important Role of m6A-Modified circRNAs in the Differentiation of Intramuscular Adipocytes in Goats Based on MeRIP Sequencing Analysis
- PMID: 36902246
- PMCID: PMC10003525
- DOI: 10.3390/ijms24054817
The Important Role of m6A-Modified circRNAs in the Differentiation of Intramuscular Adipocytes in Goats Based on MeRIP Sequencing Analysis
Abstract
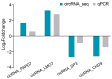
Intramuscular fat contributes to the improvement of goat meat quality. N6-Methyladenosine (m6A)-modified circular RNAs play important roles in adipocyte differentiation and metabolism. However, the mechanisms by which m6A modifies circRNA before and after differentiation of goat intramuscular adipocytes remain poorly understood. Here, we performed methylated RNA immunoprecipitation sequencing (MeRIP-seq) and circRNA sequencing (circRNA-seq) to determine the distinctions in m6A-methylated circRNAs during goat adipocyte differentiation. The profile of m6A-circRNA showed a total of 427 m6A peaks within 403 circRNAs in the intramuscular preadipocytes group, and 428 peaks within 401 circRNAs in the mature adipocytes group. Compared with the intramuscular preadipocytes group, 75 peaks within 75 circRNAs were significantly different in the mature adipocytes group. Furthermore, the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses of intramuscular preadipocytes and mature adipocytes showed that the differentially m6A-modified circRNAs were enriched in the PKG signaling pathway, endocrine and other factor-regulated calcium reabsorption, lysine degradation, etc. m6A-circRNA-miRNA-mRNA interaction networks predicted the potential m6A-circRNA regulation mechanism in different goat adipocytes. Our results indicate that there is a complicated regulatory relationship between the 12 upregulated and 7 downregulated m6A-circRNAs through 14 and 11 miRNA mediated pathways, respectively. In addition, co-analysis revealed a positive association between m6A abundance and levels of circRNA expression, such as expression levels of circRNA_0873 and circRNA_1161, which showed that m6A may play a vital role in modulating circRNA expression during goat adipocyte differentiation. These results would provide novel information for elucidating the biological functions and regulatory characteristics of m6A-circRNAs in intramuscular adipocyte differentiation and could be helpful for further molecular breeding to improve meat quality in goats.
Keywords: MeRIP-seq (m6A-seq); goat; intramuscular adipocyte; m6A-circRNAs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

