Exploration of the Shared Molecular Mechanisms between COVID-19 and Neurodegenerative Diseases through Bioinformatic Analysis
- PMID: 36902271
- PMCID: PMC10002862
- DOI: 10.3390/ijms24054839
Exploration of the Shared Molecular Mechanisms between COVID-19 and Neurodegenerative Diseases through Bioinformatic Analysis
Abstract
The COVID-19 pandemic has caused millions of deaths and remains a major public health burden worldwide. Previous studies found that a large number of COVID-19 patients and survivors developed neurological symptoms and might be at high risk of neurodegenerative diseases, such as Alzheimer's disease (AD) and Parkinson's disease (PD). We aimed to explore the shared pathways between COVID-19, AD, and PD by using bioinformatic analysis to reveal potential mechanisms, which may explain the neurological symptoms and degeneration of brain that occur in COVID-19 patients, and to provide early intervention. In this study, gene expression datasets of the frontal cortex were employed to detect common differentially expressed genes (DEGs) of COVID-19, AD, and PD. A total of 52 common DEGs were then examined using functional annotation, protein-protein interaction (PPI) construction, candidate drug identification, and regulatory network analysis. We found that the involvement of the synaptic vesicle cycle and down-regulation of synapses were shared by these three diseases, suggesting that synaptic dysfunction might contribute to the onset and progress of neurodegenerative diseases caused by COVID-19. Five hub genes and one key module were obtained from the PPI network. Moreover, 5 drugs and 42 transcription factors (TFs) were also identified on the datasets. In conclusion, the results of our study provide new insights and directions for follow-up studies of the relationship between COVID-19 and neurodegenerative diseases. The hub genes and potential drugs we identified may provide promising treatment strategies to prevent COVID-19 patients from developing these disorders.
Keywords: Alzheimer’s disease; COVID-19; Parkinson’s disease; bioinformatics; differentially expressed genes; drugs; gene ontology; hub genes; protein–protein interaction.
Conflict of interest statement
The authors have no conflict of interest to report.
Figures





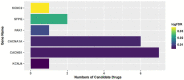

References
-
- Ceban F., Ling S., Lui L.M.W., Lee Y., Gill H., Teopiz K.M., Rodrigues N.B., Subramaniapillai M., Di Vincenzo J.D., Cao B., et al. Fatigue and cognitive impairment in Post-COVID-19 Syndrome: A systematic review and meta-analysis. Brain Behav. Immun. 2022;101:93–135. doi: 10.1016/j.bbi.2021.12.020. - DOI - PMC - PubMed
-
- Taquet M., Sillett R., Zhu L., Mendel J., Camplisson I., Dercon Q., Harrison P.J. Neurological and psychiatric risk trajectories after SARS-CoV-2 infection: An analysis of 2-year retrospective cohort studies including 1 284 437 patients. Lancet Psychiatry. 2022;9:815–827. doi: 10.1016/S2215-0366(22)00260-7. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

