DeepmRNALoc: A Novel Predictor of Eukaryotic mRNA Subcellular Localization Based on Deep Learning
- PMID: 36903531
- PMCID: PMC10005629
- DOI: 10.3390/molecules28052284
DeepmRNALoc: A Novel Predictor of Eukaryotic mRNA Subcellular Localization Based on Deep Learning
Abstract
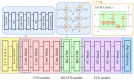
The subcellular localization of messenger RNA (mRNA) precisely controls where protein products are synthesized and where they function. However, obtaining an mRNA's subcellular localization through wet-lab experiments is time-consuming and expensive, and many existing mRNA subcellular localization prediction algorithms need to be improved. In this study, a deep neural network-based eukaryotic mRNA subcellular location prediction method, DeepmRNALoc, was proposed, utilizing a two-stage feature extraction strategy that featured bimodal information splitting and fusing for the first stage and a VGGNet-like CNN module for the second stage. The five-fold cross-validation accuracies of DeepmRNALoc in the cytoplasm, endoplasmic reticulum, extracellular region, mitochondria, and nucleus were 0.895, 0.594, 0.308, 0.944, and 0.865, respectively, demonstrating that it outperforms existing models and techniques.
Keywords: artificial intelligence; chaos-game representation; deep learning; mRNA subcellular localization.
Conflict of interest statement
The authors declare that there are no conflict of interest.
Figures



Similar articles
-
EDCLoc: a prediction model for mRNA subcellular localization using improved focal loss to address multi-label class imbalance.BMC Genomics. 2024 Dec 27;25(1):1252. doi: 10.1186/s12864-024-11173-6. BMC Genomics. 2024. PMID: 39731012 Free PMC article.
-
MSLP: mRNA subcellular localization predictor based on machine learning techniques.BMC Bioinformatics. 2023 Mar 22;24(1):109. doi: 10.1186/s12859-023-05232-0. BMC Bioinformatics. 2023. PMID: 36949389 Free PMC article.
-
Unified mRNA Subcellular Localization Predictor based on machine learning techniques.BMC Genomics. 2024 Feb 7;25(1):151. doi: 10.1186/s12864-024-10077-9. BMC Genomics. 2024. PMID: 38326777 Free PMC article.
-
A Review for Artificial Intelligence Based Protein Subcellular Localization.Biomolecules. 2024 Mar 27;14(4):409. doi: 10.3390/biom14040409. Biomolecules. 2024. PMID: 38672426 Free PMC article. Review.
-
Membrane-Coupled mRNA Trafficking in Fungi.Annu Rev Microbiol. 2015;69:265-81. doi: 10.1146/annurev-micro-091014-104242. Epub 2015 Aug 13. Annu Rev Microbiol. 2015. PMID: 26274025 Review.
Cited by
-
A review from biological mapping to computation-based subcellular localization.Mol Ther Nucleic Acids. 2023 Apr 20;32:507-521. doi: 10.1016/j.omtn.2023.04.015. eCollection 2023 Jun 13. Mol Ther Nucleic Acids. 2023. PMID: 37215152 Free PMC article. Review.
-
MSlocPRED: deep transfer learning-based identification of multi-label mRNA subcellular localization.Brief Bioinform. 2024 Sep 23;25(6):bbae504. doi: 10.1093/bib/bbae504. Brief Bioinform. 2024. PMID: 39401145 Free PMC article.
-
HIV-1 M group subtype classification using deep learning approach.Comput Biol Med. 2024 Dec;183:109218. doi: 10.1016/j.compbiomed.2024.109218. Epub 2024 Oct 5. Comput Biol Med. 2024. PMID: 39369547
-
lncRNA localization and feature interpretability analysis.Mol Ther Nucleic Acids. 2024 Dec 12;36(1):102425. doi: 10.1016/j.omtn.2024.102425. eCollection 2025 Mar 11. Mol Ther Nucleic Acids. 2024. PMID: 39926317 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

