Design of Novel Phosphatidylinositol 3-Kinase Inhibitors for Non-Hodgkin's Lymphoma: Molecular Docking, Molecular Dynamics, and Density Functional Theory Studies on Gold Nanoparticles
- PMID: 36903539
- PMCID: PMC10005307
- DOI: 10.3390/molecules28052289
Design of Novel Phosphatidylinositol 3-Kinase Inhibitors for Non-Hodgkin's Lymphoma: Molecular Docking, Molecular Dynamics, and Density Functional Theory Studies on Gold Nanoparticles
Abstract
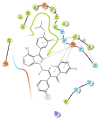
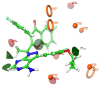
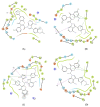
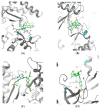
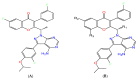
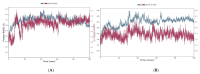
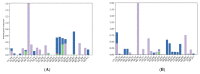
Non-Hodgkin's lymphomas are a diverse collection of lymphoproliferative cancers that are much less predictable than Hodgkin's lymphomas with a far greater tendency to metastasize to extranodal sites. A quarter of non-Hodgkin's lymphoma cases develop at extranodal sites and the majority of them involve nodal and extranodal sites. The most common subtypes include follicular lymphoma, chronic/small lymphocytic leukaemia, mantel cell lymphoma, and marginal zone lymphoma. Umbralisib is one of the latest PI3Kδ inhibitors in clinical trials for several hematologic cancer indications. In this study, new umbralisib analogues were designed and docked to the active site of PI3Kδ, the main target of the phosphoinositol-3-kinase/Akt/mammalian target of the rapamycin pathway (PI3K/AKT/mTOR). This study resulted in eleven candidates, with strong binding to PI3Kδ with a docking score between -7.66 and -8.42 Kcal/mol. The docking analysis of ligand-receptor interactions between umbralisib analogues bound to PI3K showed that their interactions were mainly controlled by hydrophobic interactions and, to a lesser extent, by hydrogen bonding. In addition, the MM-GBSA binding free energy was calculated. Analogue 306 showed the highest free energy of binding with -52.22 Kcal/mol. To identify the structural changes and the complexes' stability of proposed ligands, molecular dynamic simulation was used. Based on this research finding, the best-designed analogue, analogue 306, formed a stable ligand-protein complex. In addition, pharmacokinetics and toxicity analysis using the QikProp tool demonstrated that analogue 306 had good absorption, distribution, metabolism, and excretion properties. Additionally, it has a promising predicted profile in immune toxicity, carcinogenicity, and cytotoxicity. In addition, analogue 306 had stable interactions with gold nanoparticles that have been studied using density functional theory calculations. The best interaction with gold was observed at the oxygen atom number 5 with -29.42 Kcal/mol. Further in vitro and in vivo investigations are recommended to be carried out to verify the anticancer activity of this analogue.
Keywords: cancer; drug discovery; gold nanoparticles; health and wellbeing; molecular docking; molecular dynamics; pi3k; umbralisib analogues.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Identification of novel PI3Kδ inhibitors by docking, ADMET prediction and molecular dynamics simulations.Comput Biol Chem. 2019 Feb;78:190-204. doi: 10.1016/j.compbiolchem.2018.12.002. Epub 2018 Dec 7. Comput Biol Chem. 2019. PMID: 30557817
-
Umbralisib, a novel PI3Kδ and casein kinase-1ε inhibitor, in relapsed or refractory chronic lymphocytic leukaemia and lymphoma: an open-label, phase 1, dose-escalation, first-in-human study.Lancet Oncol. 2018 Apr;19(4):486-496. doi: 10.1016/S1470-2045(18)30082-2. Epub 2018 Feb 20. Lancet Oncol. 2018. PMID: 29475723 Clinical Trial.
-
Rituximab: a review of its use in non-Hodgkin's lymphoma and chronic lymphocytic leukaemia.Drugs. 2003;63(8):803-43. doi: 10.2165/00003495-200363080-00005. Drugs. 2003. PMID: 12662126 Review.
-
Network Pharmacology and In silico Elucidation of Phytochemicals Extracted from Ajwa Dates (Phoenix dactylifera L.) to Inhibit Akt and PI3K Causing Triple Negative Breast Cancer (TNBC).Curr Pharm Des. 2025;31(10):774-796. doi: 10.2174/0113816128348876241017101729. Curr Pharm Des. 2025. PMID: 39698883
-
Umbralisib: First Approval.Drugs. 2021 May;81(7):857-866. doi: 10.1007/s40265-021-01504-2. Drugs. 2021. PMID: 33797740 Review.
Cited by
-
Screening Enamine Fragments Library in the Quest for Novel SOS2 Inhibitors: Pharmacophore Modelling, Molecular Docking, MMGBSA Calculations, and MD Simulation.J Pharm Bioallied Sci. 2025 Jun;17(Suppl 2):S1894-S1899. doi: 10.4103/jpbs.jpbs_534_25. Epub 2025 Jun 18. J Pharm Bioallied Sci. 2025. PMID: 40655702 Free PMC article.
References
-
- Binder A.F., Brody J.D. Non-Hodgkin Lymphoma. Oncology. 2021:342–353. doi: 10.1002/9781119189596.ch30. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

