Genome-Wide Identification and Expression Profiling of Glutathione S-Transferase Gene Family in Foxtail Millet (Setaria italica L.)
- PMID: 36904001
- PMCID: PMC10005783
- DOI: 10.3390/plants12051138
Genome-Wide Identification and Expression Profiling of Glutathione S-Transferase Gene Family in Foxtail Millet (Setaria italica L.)
Abstract
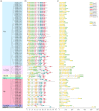
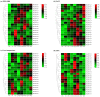
Glutathione S-transferases (GSTs) are a critical superfamily of multifunctional enzymes in plants. As a ligand or binding protein, GSTs regulate plant growth and development and detoxification. Foxtail millet (Setaria italica (L.) P. Beauv) could respond to abiotic stresses through a highly complex multi-gene regulatory network in which the GST family is also involved. However, GST genes have been scarcely studied in foxtail millet. Genome-wide identification and expression characteristics analysis of the foxtail millet GST gene family were conducted by biological information technology. The results showed that 73 GST genes (SiGSTs) were identified in the foxtail millet genome and were divided into seven classes. The chromosome localization results showed uneven distribution of GSTs on the seven chromosomes. There were 30 tandem duplication gene pairs belonging to 11 clusters. Only one pair of SiGSTU1 and SiGSTU23 were identified as fragment duplication genes. A total of ten conserved motifs were identified in the GST family of foxtail millet. The gene structure of SiGSTs is relatively conservative, but the number and length of exons of each gene are still different. The cis-acting elements in the promoter region of 73 SiGST genes showed that 94.5% of SiGST genes possessed defense and stress-responsive elements. The expression profiles of 37 SiGST genes covering 21 tissues suggested that most SiGST genes were expressed in multiple organs and were highly expressed in roots and leaves. By qPCR analysis, we found that 21 SiGST genes were responsive to abiotic stresses and abscisic acid (ABA). Taken together, this study provides a theoretical basis for identifying foxtail millet GST family information and improving their responses to different stresses.
Keywords: expression analysis; foxtail millet; glutathione S-transferase (GSTs); stress response.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Abdul Kayum M., Nath U.K., Park J.I., Biswas M.K., Choi E.K., Song J.Y., Kim H.T., Nou I.S. Genome-Wide Identification, Characterization, and Expression Profiling of Glutathione S-Transferase (GST) Family in Pumpkin Reveals Likely Role in Cold-Stress Tolerance. Genes. 2018;9:84. doi: 10.3390/genes9020084. - DOI - PMC - PubMed
-
- Sasan M., Maryam E., Fateme M., Maryam S., Babak S., Hassan M. Plant glutathione S-transferase classification, structure and evolution. Afr. J. Biochem. Res. 2011;10:8160–8165.
Grants and funding
- YJHZKF2104 and 202003-5/State Key Laboratory of Sustainable Dryland Agriculture (in preparation), Shanxi Agricultural University
- 20210302124699/Shanxi Province Basic Research Program Project
- 2021BQ20/Doctoral Research Start Project of Shanxi Agricultural University
- SXBYKY2021044/Shanxi province doctoral graduates and postdoctoral researchers to Work Award Fund Research Project
- CARS-06-14.5-A28/China Agriculture Research System of MOF and MARA
LinkOut - more resources
Full Text Sources
Research Materials

