A cryptic natural variant allele of BYPASS2 suppresses the bypass1 mutant phenotype
- PMID: 36905371
- PMCID: PMC10231379
- DOI: 10.1093/plphys/kiad124
A cryptic natural variant allele of BYPASS2 suppresses the bypass1 mutant phenotype
Abstract
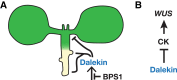
The Arabidopsis (Arabidopsis thaliana) BYPASS1 (BPS1) gene encodes a protein with no functionally characterized domains, and loss-of-function mutants (e.g. bps1-2 in Col-0) present a severe growth arrest phenotype that is evoked by a root-derived graft-transmissible small molecule that we call dalekin. The root-to-shoot nature of dalekin signaling suggests it could be an endogenous signaling molecule. Here, we report a natural variant screen that allowed us to identify enhancers and suppressors of the bps1-2 mutant phenotype (in Col-0). We identified a strong semi-dominant suppressor in the Apost-1 accession that largely restored shoot development in bps1 and yet continued to overproduce dalekin. Using bulked segregant analysis and allele-specific transgenic complementation, we showed that the suppressor is the Apost-1 allele of a BPS1 paralog, BYPASS2 (BPS2). BPS2 is one of four members of the BPS gene family in Arabidopsis, and phylogenetic analysis demonstrated that the BPS family is conserved in land plants and the four Arabidopsis paralogs are retained duplicates from whole genome duplications. The strong conservation of BPS1 and paralogous proteins throughout land plants, and the similar functions of paralogs in Arabidopsis, suggests that dalekin signaling might be retained across land plants.
© American Society of Plant Biologists 2023. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
Conflict of interest statement. L.E.S. and A.J.C. have a confict of interest related to experimental outcomes described in this publication.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

