USP22 upregulates ZEB1-mediated VEGFA transcription in hepatocellular carcinoma
- PMID: 36906615
- PMCID: PMC10008583
- DOI: 10.1038/s41419-023-05699-y
USP22 upregulates ZEB1-mediated VEGFA transcription in hepatocellular carcinoma
Abstract
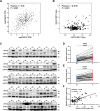
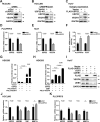
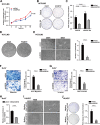
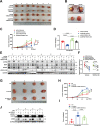
Hepatocellular carcinoma (HCC) is a common solid tumor with high rate of recurrence and mortality. Anti-angiogenesis drugs have been used for the therapy of HCC. However, anti-angiogenic drug resistance commonly occurs during HCC treatment. Thus, identification of a novel VEGFA regulator would be better understanding for HCC progression and anti-angiogenic therapy resistance. Ubiquitin specific protease 22 (USP22) as a deubiquitinating enzyme, participates in a variety of biological processes in numerous tumors. While the molecular mechanism underlying the effects of USP22 on angiogenesis is still needed to be clarified. Here, our results demonstrated that USP22 acts as a co-activator of VEGFA transcription. Importantly, USP22 is involved in maintenance of ZEB1 stability via its deubiquitinase activity. USP22 was recruited to ZEB1-binding elements on the promoter of VEGFA, thereby altering histone H2Bub levels, to enhance ZEB1-mediated VEGFA transcription. USP22 depletion decreased cell proliferation, migration, Vascular Mimicry (VM) formation, and angiogenesis. Furthermore, we provided the evidence to show that knockdown of USP22 inhibited HCC growth in tumor-bearing nude mice. In addition, the expression of USP22 is positively correlated with that of ZEB1 in clinical HCC samples. Our findings suggest that USP22 participates in the promotion of HCC progression, if not all, at least partially via up-regulation of VEGFA transcription, providing a novel therapeutic target for anti-angiogenic drug resistance in HCC.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

