Molecular Basis and Genetic Modifiers of Thalassemia
- PMID: 36907603
- PMCID: PMC10122828
- DOI: 10.1016/j.hoc.2022.12.001
Molecular Basis and Genetic Modifiers of Thalassemia
Abstract
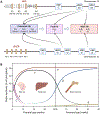
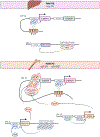
Thalassemia syndromes are common monogenic disorders and represent a significant health issue worldwide. In this review, the authors elaborate on fundamental genetic knowledge about thalassemias, including the structure and location of globin genes, the production of hemoglobin during development, the molecular lesions causing α-, β-, and other thalassemia syndromes, the genotype-phenotype correlation, and the genetic modifiers of these conditions. In addition, they briefly discuss the molecular techniques applied for diagnosis and innovative cell and gene therapy strategies to cure these conditions.
Keywords: Genetic modifiers; Genotype-phenotype; Globin genes; Hemoglobin; Hemoglobin switch; Molecular genetics; Thalassemia.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Disclosure The authors have nothing to disclose.
Figures




References
-
- Kattamis A, Kwiatkowski JL, Aydinok Y. Thalassaemia. Lancet. 2022;399(10343):2310–2324. - PubMed
-
- Weatherall DJ. Genetic variation and susceptibility to infection: the red cell and malaria. Br J Haematol. 2008;141(3):276–286. - PubMed
-
- Weatherall DJ. The Evolving Spectrum of the Epidemiology of Thalassemia. Hematol Oncol Clin North Am. 2018;32(2):165–175. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

