This is a preprint.
Smaug regulates germ plasm synthesis and primordial germ cell number in Drosophila embryos by repressing the oskar and bruno 1 mRNAs
- PMID: 36909513
- PMCID: PMC10002672
- DOI: 10.1101/2023.02.27.530189
Smaug regulates germ plasm synthesis and primordial germ cell number in Drosophila embryos by repressing the oskar and bruno 1 mRNAs
Update in
-
Smaug regulates germ plasm assembly and primordial germ cell number in Drosophila embryos.Sci Adv. 2024 Apr 12;10(15):eadg7894. doi: 10.1126/sciadv.adg7894. Epub 2024 Apr 12. Sci Adv. 2024. PMID: 38608012 Free PMC article.
Abstract
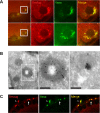
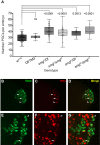
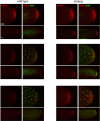
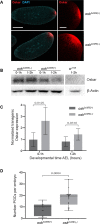
During Drosophila oogenesis, the Oskar (OSK) RNA-binding protein (RBP) determines the amount of germ plasm that assembles at the posterior pole of the oocyte. Here we identify the mechanisms that regulate the osk mRNA in the early embryo. We show that the Smaug (SMG) RBP is transported into the germ plasm of the early embryo where it accumulates in the germ granules. SMG binds to and represses translation of the osk mRNA itself as well as the bruno 1 (bru1) mRNA, which encodes an RBP that we show promotes germ plasm production. Loss of SMG or mutation of SMG's binding sites in the osk or bru1 mRNAs results in ectopic translation of these transcripts in the germ plasm and excess PGCs. SMG therefore triggers a post-transcriptional regulatory pathway that attenuates germ plasm synthesis in embryos, thus modulating the number of PGCs.
Conflict of interest statement
Competing interests: All authors declare they have no competing interests.
Figures

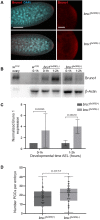

References
-
- A. P. Mahowald, Assembly of the Drosophila germ plasm. Int Rev Cytol 203, 187–213 (2001). - PubMed
-
- Houston D. W., King M. L., Germ plasm and molecular determinants of germ cell fate. Curr Top Dev Biol 50, 155–181 (2000). - PubMed
-
- Rongo C. et al., Germ plasm assembly and germ cell migration in Drosophila. Cold Spring Harb Symp Quant Biol 62, 1–11 (1997). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
