This is a preprint.
Active maintenance of CD8+ T cell naïvety through regulation of global genome architecture
- PMID: 36909629
- PMCID: PMC10002700
- DOI: 10.1101/2023.02.26.530139
Active maintenance of CD8+ T cell naïvety through regulation of global genome architecture
Update in
-
Active maintenance of CD8+ T cell naivety through regulation of global genome architecture.Cell Rep. 2023 Oct 31;42(10):113301. doi: 10.1016/j.celrep.2023.113301. Epub 2023 Oct 19. Cell Rep. 2023. PMID: 37858463 Free PMC article.
Abstract
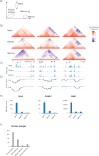
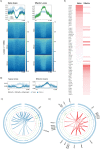
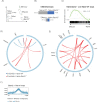
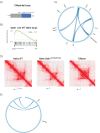
The differentiation of naïve CD8+ cytotoxic T lymphocytes (CTLs) into effector and memory states results in large scale changes in transcriptional and phenotypic profiles. Little is known about how large-scale changes in genome organisation reflect or underpin these transcriptional programs. We utilised Hi-C to map changes in the spatial organisation of long-range genome contacts within naïve, effector and memory virus-specific CD8+ T cells. We observed that the architecture of the naive CD8+ T cell genome was distinct from effector and memory genome configurations with extensive changes within discrete functional chromatin domains. However, deletion of the BACH2 or SATB1 transcription factors was sufficient to remodel the naïve chromatin architecture and engage transcriptional programs characteristic of differentiated cells. This suggests that the chromatin architecture within naïve CD8+ T cells is preconfigured to undergo autonomous remodelling upon activation, with key transcription factors restraining differentiation by actively enforcing the unique naïve chromatin state.
Conflict of interest statement
DECLARATION OF INTERESTS S.J.T. is a member of the scientific advisory board for Medicago, Inc., QC, Canada. No funding from Medicago was provided for this work. A.W.G. is a member of the scientific advisory board of ArsenalBio.
Figures



References
-
- Agarwal S., and Rao A. (1998). Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity 9, 765–775. - PubMed
-
- Avni O., Lee D., Macian F., Szabo S.J., Glimcher L.H., and Rao A. (2002). T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat Immunol 3, 643–651. - PubMed
-
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., and Zhao K. (2007). High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
