Single-cell transcriptomic heterogeneity between conduit and resistance mesenteric arteries in rats
- PMID: 36912534
- PMCID: PMC10085562
- DOI: 10.1152/physiolgenomics.00126.2022
Single-cell transcriptomic heterogeneity between conduit and resistance mesenteric arteries in rats
Abstract
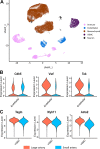
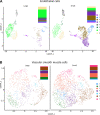
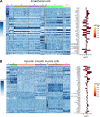
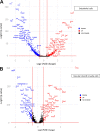
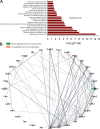
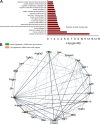
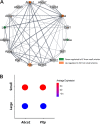
The endothelium contains morphologically similar cells throughout the vasculature, but individual cells along the length of a single vascular tree or in different regional circulations function dissimilarly. When observations made in large arteries are extrapolated to explain the function of endothelial cells (ECs) in the resistance vasculature, only a fraction of these observations are consistent between artery sizes. To what extent endothelial (EC) and vascular smooth muscle cells (VSMCs) from different arteriolar segments of the same tissue differ phenotypically at the single-cell level remains unknown. Therefore, single-cell RNA-seq (10x Genomics) was performed using a 10X Genomics Chromium system. Cells were enzymatically digested from large (>300 µm) and small (<150 µm) mesenteric arteries from nine adult male Sprague-Dawley rats, pooled to create six samples (3 rats/sample, 3 samples/group). After normalized integration, the dataset was scaled before unsupervised cell clustering and cluster visualization using UMAP plots. Differential gene expression analysis allowed us to infer the biological identity of different clusters. Our analysis revealed 630 and 641 differentially expressed genes (DEGs) between conduit and resistance arteries for ECs and VSMCs, respectively. Gene ontology analysis (GO-Biological Processes, GOBP) of scRNA-seq data discovered 562 and 270 pathways for ECs and VSMCs, respectively, that differed between large and small arteries. We identified eight and seven unique ECs and VSMCs subpopulations, respectively, with DEGs and pathways identified for each cluster. These results and this dataset allow the discovery and support of novel hypotheses needed to identify mechanisms that determine the phenotypic heterogeneity between conduit and resistance arteries.
Keywords: endothelial; heterogeneity; mesenteric; single-cell RNAseq; smooth muscle.
Conflict of interest statement
No conflicts of interest, financial or otherwise, are declared by the authors.
Figures

References
-
- Schuliga M. Smooth muscle and extracellular matrix interactions in health and disease. In: Muscle Cell and Tissue, edited by Sakuma K. London: IntechOpen, 2015, p. 486. doi: 10.5772/60403. - DOI
-
- Jiang J, Zheng JP, Li Y, Gan Z, Jiang Y, Huang D, Li H, Liu Z, Ke Y. Differential contribution of endothelium-derived relaxing factors to vascular reactivity in conduit and resistance arteries from normotensive and hypertensive rats. Clin Exp Hypertens 38: 393–398, 2016. doi: 10.3109/10641963.2016.1148155. - DOI - PubMed
-
- Vanlandewijck M, He L, Mäe MA, Andrae J, Ando K, Del Gaudio F, Nahar K, Lebouvier T, Laviña B, Gouveia L, Sun Y, Raschperger E, Räsänen M, Zarb Y, Mochizuki N, Keller A, Lendahl U, Betsholtz C. A molecular atlas of cell types and zonation in the brain vasculature. Nature 554: 475–480, 2018. [Erratum in Nature 560: E3, 2018]. doi: 10.1038/nature25739. - DOI - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

