Analysis of companion cell and phloem metabolism using a transcriptome-guided model of Arabidopsis metabolism
- PMID: 36913519
- PMCID: PMC10231466
- DOI: 10.1093/plphys/kiad154
Analysis of companion cell and phloem metabolism using a transcriptome-guided model of Arabidopsis metabolism
Abstract
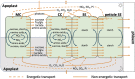
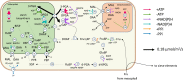
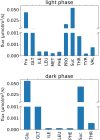
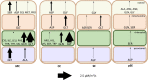
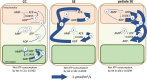
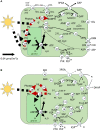
Companion cells and sieve elements play an essential role in vascular plants, and yet the details of the metabolism that underpins their function remain largely unknown. Here, we construct a tissue-scale flux balance analysis (FBA) model to describe the metabolism of phloem loading in a mature Arabidopsis (Arabidopsis thaliana) leaf. We explore the potential metabolic interactions between mesophyll cells, companion cells, and sieve elements based on the current understanding of the physiology of phloem tissue and through the use of cell type-specific transcriptome data as a weighting in our model. We find that companion cell chloroplasts likely play a very different role to mesophyll chloroplasts. Our model suggests that, rather than carbon capture, the most crucial function of companion cell chloroplasts is to provide photosynthetically generated ATP to the cytosol. Additionally, our model predicts that the metabolites imported into the companion cell are not necessarily the same metabolites that are exported in phloem sap; phloem loading is more efficient if certain amino acids are synthesized in the phloem tissue. Surprisingly, in our model predictions, the proton-pumping pyrophosphatase (H+-PPiase) is a more efficient contributor to the energization of the companion cell plasma membrane than the H+-ATPase.
© The Author(s) 2023. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures

References
-
- Ambasht PK, Kayastha AM. Plant pyruvate kinase. Biol Plant. 2002:45(1):1–10. 10.1023/A:1015173724712 - DOI
-
- Bao A-K, Wang S-M, Wu G-Q, Xi J-J, Zhang J-L, Wang C-M. Overexpression of the Arabidopsis H+-PPase enhanced resistance to salt and drought stress in transgenic alfalfa (Medicago sativa L.). Plant Sci. 2009:176(2):232–240. 10.1016/j.plantsci.2008.10.009 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

