Rgs1 is a regulator of effector gene expression during plant infection by the rice blast fungus Magnaporthe oryzae
- PMID: 36913579
- PMCID: PMC10041150
- DOI: 10.1073/pnas.2301358120
Rgs1 is a regulator of effector gene expression during plant infection by the rice blast fungus Magnaporthe oryzae
Abstract
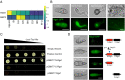
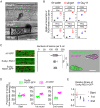
To cause rice blast disease, the filamentous fungus Magnaporthe oryzae secretes a battery of effector proteins into host plant tissue to facilitate infection. Effector-encoding genes are expressed only during plant infection and show very low expression during other developmental stages. How effector gene expression is regulated in such a precise manner during invasive growth by M. oryzae is not known. Here, we report a forward-genetic screen to identify regulators of effector gene expression, based on the selection of mutants that show constitutive effector gene expression. Using this simple screen, we identify Rgs1, a regulator of G-protein signaling (RGS) protein that is necessary for appressorium development, as a novel transcriptional regulator of effector gene expression, which acts prior to plant infection. We show that an N-terminal domain of Rgs1, possessing transactivation activity, is required for effector gene regulation and acts in an RGS-independent manner. Rgs1 controls the expression of at least 60 temporally coregulated effector genes, preventing their transcription during the prepenetration stage of development prior to plant infection. A regulator of appressorium morphogenesis is therefore also required for the orchestration of pathogen gene expression required for invasive growth by M. oryzae during plant infection.
Keywords: effectors; gene expression; plant pathogen; rice blast.
Conflict of interest statement
The authors declare no competing interest.
Figures



References
-
- Lo Presti L., et al. , Fungal effectors and plant susceptibility. Annu. Rev. Plant Biol. 66, 513–545 (2015). - PubMed
-
- Kamoun S., A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 44, 41–60 (2006). - PubMed
-
- Skamnioti P., Gurr S. J., Against the grain: Safeguarding rice from rice blast disease. Trends Biotechnol. 27, 141–150 (2009). - PubMed
-
- Jeon J., et al. , Transcriptome profiling of the rice blast fungus Magnaporthe oryzae and its host oryza sativa during infection. Mol. Plant Microb. Interact. 33, 141–144 (2019). - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

