An anti-HER2 biparatopic antibody that induces unique HER2 clustering and complement-dependent cytotoxicity
- PMID: 36914633
- PMCID: PMC10011572
- DOI: 10.1038/s41467-023-37029-3
An anti-HER2 biparatopic antibody that induces unique HER2 clustering and complement-dependent cytotoxicity
Abstract
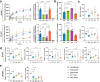
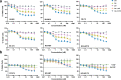
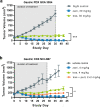
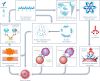
Human epidermal growth factor receptor 2 (HER2) is a receptor tyrosine kinase that plays an oncogenic role in breast, gastric and other solid tumors. However, anti-HER2 therapies are only currently approved for the treatment of breast and gastric/gastric esophageal junction cancers and treatment resistance remains a problem. Here, we engineer an anti-HER2 IgG1 bispecific, biparatopic antibody (Ab), zanidatamab, with unique and enhanced functionalities compared to both trastuzumab and the combination of trastuzumab plus pertuzumab (tras + pert). Zanidatamab binds adjacent HER2 molecules in trans and initiates distinct HER2 reorganization, as shown by polarized cell surface HER2 caps and large HER2 clusters, not observed with trastuzumab or tras + pert. Moreover, zanidatamab, but not trastuzumab nor tras + pert, elicit potent complement-dependent cytotoxicity (CDC) against high HER2-expressing tumor cells in vitro. Zanidatamab also mediates HER2 internalization and downregulation, inhibition of both cell signaling and tumor growth, antibody-dependent cellular cytotoxicity (ADCC) and phagocytosis (ADCP), and also shows superior in vivo antitumor activity compared to tras + pert in a HER2-expressing xenograft model. Collectively, we show that zanidatamab has multiple and distinct mechanisms of action derived from the structural effects of biparatopic HER2 engagement.
© 2023. The Author(s).
Conflict of interest statement
Zymeworks Inc. has filed patent applications relating to zanidatamab: PCT/CA2014/051140 (N.E.W., G.N., G.W., S.D., E.E.C., and M.S. are listed as inventors) and PCT/CA2015/051238 (N.E.W., G.N., G.W. are listed as inventors). These applications have issued in several jurisdictions, including the United States [US 10,000,576; US 10,947,319; US 11,325,981] and Japan [JP 6727379; JP 6817064; JP 6872482]. The other authors declare no competing interests.
Figures
References
-
- Bang YJ, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–697. doi: 10.1016/S0140-6736(10)61121-X. - DOI - PubMed
-
- HERCEPTIN® (trastuzumab). Prescribing Information (Genentech, Inc., South San Francisco, CA, 2018).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

