Age-associated microenvironmental changes highlight the role of PDGF-C in ER+ breast cancer metastatic relapse
- PMID: 36914817
- PMCID: PMC10132974
- DOI: 10.1038/s43018-023-00525-y
Age-associated microenvironmental changes highlight the role of PDGF-C in ER+ breast cancer metastatic relapse
Abstract
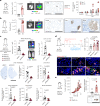
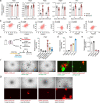
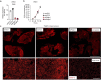
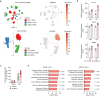
Patients with estrogen receptor (ER)-positive breast cancer are at risk of metastatic relapse for decades after primary tumor resection and treatment, a consequence of dormant disseminated tumor cells (DTCs) reawakening at secondary sites. Here we use syngeneic ER+ mouse models in which DTCs display a dormant phenotype in young mice but accelerated metastatic outgrowth in an aged or fibrotic microenvironment. In young mice, low-level Pdgfc expression by ER+ DTCs is required for their maintenance in secondary sites but is insufficient to support development of macrometastases. By contrast, the platelet-derived growth factor (PDGF)-Chi environment of aging or fibrotic lungs promotes DTC proliferation and upregulates tumor cell Pdgfc expression stimulating further stromal activation, events that can be blocked by pharmacological inhibition of PDGFRα or with a PDGF-C-blocking antibody. These results highlight the role of the changing microenvironment in regulating DTC outgrowth and the opportunity to target PDGF-C signaling to limit metastatic relapse in ER+ breast cancer.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures













Comment in
-
The aging lung reawakens dormant tumor cells.Nat Cancer. 2023 Apr;4(4):442-443. doi: 10.1038/s43018-023-00537-8. Nat Cancer. 2023. PMID: 37106111 No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

