Bipotent transitional liver progenitor cells contribute to liver regeneration
- PMID: 36914834
- PMCID: PMC10101857
- DOI: 10.1038/s41588-023-01335-9
Bipotent transitional liver progenitor cells contribute to liver regeneration
Erratum in
-
Author Correction: Bipotent transitional liver progenitor cells contribute to liver regeneration.Nat Genet. 2023 May;55(5):892. doi: 10.1038/s41588-023-01393-z. Nat Genet. 2023. PMID: 37072629 Free PMC article. No abstract available.
Abstract
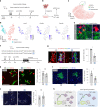
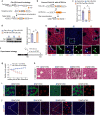
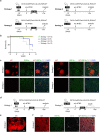
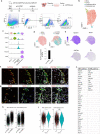
Following severe liver injury, when hepatocyte-mediated regeneration is impaired, biliary epithelial cells (BECs) can transdifferentiate into functional hepatocytes. However, the subset of BECs with such facultative tissue stem cell potential, as well as the mechanisms enabling transdifferentiation, remains elusive. Here we identify a transitional liver progenitor cell (TLPC), which originates from BECs and differentiates into hepatocytes during regeneration from severe liver injury. By applying a dual genetic lineage tracing approach, we specifically labeled TLPCs and found that they are bipotent, as they either differentiate into hepatocytes or re-adopt BEC fate. Mechanistically, Notch and Wnt/β-catenin signaling orchestrate BEC-to-TLPC and TLPC-to-hepatocyte conversions, respectively. Together, our study provides functional and mechanistic insights into transdifferentiation-assisted liver regeneration.
© 2023. The Author(s).
Conflict of interest statement
M.P. and J.S.T. are shareholders of and employed by Novartis Pharma AG but declare no conflict of interest associated with this work. The remaining authors declare no competing interests.
Figures













References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

