Preneoplastic stromal cells promote BRCA1-mediated breast tumorigenesis
- PMID: 36914836
- PMCID: PMC10655552
- DOI: 10.1038/s41588-023-01298-x
Preneoplastic stromal cells promote BRCA1-mediated breast tumorigenesis
Abstract
Women with germline BRCA1 mutations (BRCA1+/mut) have increased risk for hereditary breast cancer. Cancer initiation in BRCA1+/mut is associated with premalignant changes in breast epithelium; however, the role of the epithelium-associated stromal niche during BRCA1-driven tumor initiation remains unclear. Here we show that the premalignant stromal niche promotes epithelial proliferation and mutant BRCA1-driven tumorigenesis in trans. Using single-cell RNA sequencing analysis of human preneoplastic BRCA1+/mut and noncarrier breast tissues, we show distinct changes in epithelial homeostasis including increased proliferation and expansion of basal-luminal intermediate progenitor cells. Additionally, BRCA1+/mut stromal cells show increased expression of pro-proliferative paracrine signals. In particular, we identify pre-cancer-associated fibroblasts (pre-CAFs) that produce protumorigenic factors including matrix metalloproteinase 3 (MMP3), which promotes BRCA1-driven tumorigenesis in vivo. Together, our findings demonstrate that precancerous stroma in BRCA1+/mut may elevate breast cancer risk through the promotion of epithelial proliferation and an accumulation of luminal progenitor cells with altered differentiation.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
All the other authors declare no competing interests.
Figures










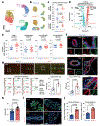

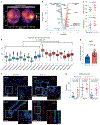
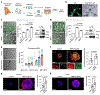


Comment in
-
Stromal cells drive tumorigenesis in BRCA1 mutation carriers.Nat Rev Cancer. 2023 Jun;23(6):349. doi: 10.1038/s41568-023-00580-8. Nat Rev Cancer. 2023. PMID: 37101004 No abstract available.
References
-
- Wooster R & Weber BL Breast and ovarian cancer. N. Engl. J. Med 348, 2339–2347 (2003). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

