The genetic basis of endometriosis and comorbidity with other pain and inflammatory conditions
- PMID: 36914876
- PMCID: PMC10042257
- DOI: 10.1038/s41588-023-01323-z
The genetic basis of endometriosis and comorbidity with other pain and inflammatory conditions
Abstract
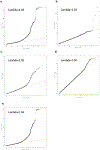
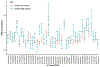
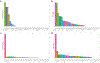
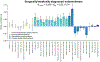
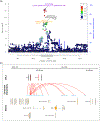
Endometriosis is a common condition associated with debilitating pelvic pain and infertility. A genome-wide association study meta-analysis, including 60,674 cases and 701,926 controls of European and East Asian descent, identified 42 genome-wide significant loci comprising 49 distinct association signals. Effect sizes were largest for stage 3/4 disease, driven by ovarian endometriosis. Identified signals explained up to 5.01% of disease variance and regulated expression or methylation of genes in endometrium and blood, many of which were associated with pain perception/maintenance (SRP14/BMF, GDAP1, MLLT10, BSN and NGF). We observed significant genetic correlations between endometriosis and 11 pain conditions, including migraine, back and multisite chronic pain (MCP), as well as inflammatory conditions, including asthma and osteoarthritis. Multitrait genetic analyses identified substantial sharing of variants associated with endometriosis and MCP/migraine. Targeted investigations of genetically regulated mechanisms shared between endometriosis and other pain conditions are needed to aid the development of new treatments and facilitate early symptomatic intervention.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing Interests
L.S., G.T., K.S. and V.S. are employees of deCODE Genetics/Amgen. G.G., C.C. and P.Y.B. are employees of Celmatix Inc., and hold stock or stock options in Celmatix Inc. P.F. and J.Y.T. are employees of 23andMe Inc. O.H. serves occasionally on advisory boards for Bayer AG, Gedeon Richter, HRA Pharma, and Vifor Pharma, and has lectured at educational events for Bayer AG, Gedeon Richter, and Sandoz AG. C.M.L. receives grant support from Bayer AG, Novo Nordisk and her husband works for Vertex. J.R. spouse receives salary and stock options from Merck & Co. K.V. receives research funding from Bayer Healthcare and Honorarium for Consultancy and Lectures from Bayer Healthcare, Grunenthal GmBH, AbbVie and Eli Lilly. T.D. is employee of Merck Healthcare KGaA, Darmstadt, Germany. T. S. is the co-founder and shareholder of Zoe Global Ltd. P.A.W.R. is an advisory board member for Bayer AG. O.U. lectured at educational events for Exeltis and Merck. K.L.T. report an endometriosis grant from Aspira which is not relevant to the submitted work. K.T.Z. and C.M.B. reported grants from Bayer AG, AbbVie Inc, Volition Rx, MDNA Life Sciences, Roche Diagnostics Inc. outside the submitted work. The remaining authors declare no competing interests.
Figures


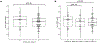



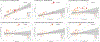

References
-
- Zondervan KT, Becker CM & Missmer SA Endometriosis. N Engl J Med 382, 1244–1256 (2020). - PubMed
-
- ASRM. Revised American Society for Reproductive Medicine classification of endometriosis: 1996. Fertil Steril 67, 817–21 (1997). - PubMed
-
- Saha R et al. Heritability of endometriosis. Fertil Steril 104, 947–952 (2015). - PubMed
-
- Treloar SA, O’Connor DT, O’Connor VM & Martin NG Genetic influences on endometriosis in an Australian twin sample. sueT@qimr.edu.au. Fertil Steril 71, 701–10 (1999). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

