Infection with wild-type SARS-CoV-2 elicits broadly neutralizing and protective antibodies against omicron subvariants
- PMID: 36914890
- PMCID: PMC10063446
- DOI: 10.1038/s41590-023-01449-6
Infection with wild-type SARS-CoV-2 elicits broadly neutralizing and protective antibodies against omicron subvariants
Abstract
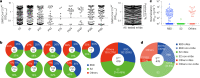
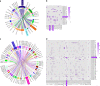
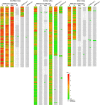
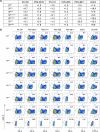
The omicron variants of SARS-CoV-2 have substantial ability to escape infection- and vaccine-elicited antibody immunity. Here, we investigated the extent of such escape in nine convalescent patients infected with the wild-type SARS-CoV-2 during the first wave of the pandemic. Among the total of 476 monoclonal antibodies (mAbs) isolated from peripheral memory B cells, we identified seven mAbs with broad neutralizing activity to all variants tested, including various omicron subvariants. Biochemical and structural analysis indicated the majority of these mAbs bound to the receptor-binding domain, mimicked the receptor ACE2 and were able to accommodate or inadvertently improve recognition of omicron substitutions. Passive delivery of representative antibodies protected K18-hACE2 mice from infection with omicron and beta SARS-CoV-2. A deeper understanding of how the memory B cells that produce these antibodies could be selectively boosted or recalled can augment antibody immunity against SARS-CoV-2 variants.
© 2023. The Author(s).
Conflict of interest statement
Patent applications have been filed that cover some of the antibodies presented here (patent application nos. 202310046052.1, 202310046053.6, 202310046054.0, 202310046051.7, 202310046056.X, 202310107070.6, 202310130257.8, 202310050519.X, 202310118826.7, 202310050250.5, 202310050475.0 and 202310050170.X). L.Z., Z.Z., Q.Z., B.J., P.C., B.Z. and X.S. are the inventors. The other authors declare no competing interests.
Figures











References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

