Distinct in vitro and in vivo neutralization profiles of monoclonal antibodies elicited by the receptor binding domain of the ancestral SARS-CoV-2
- PMID: 36916782
- PMCID: PMC10189799
- DOI: 10.1002/jmv.28673
Distinct in vitro and in vivo neutralization profiles of monoclonal antibodies elicited by the receptor binding domain of the ancestral SARS-CoV-2
Abstract
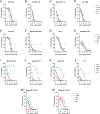
Broadly neutralizing antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants are sought to curb coronavirus disease 2019 (COVID-19) infections. Here we produced and characterized a set of mouse monoclonal antibodies (mAbs) specific for the ancestral SARS-CoV-2 receptor binding domain (RBD). Two of them, 17A7 and 17B10, were highly potent in microneutralization assay with 50% inhibitory concentration (IC50 ) ≤135 ng/mL against infectious SARS-CoV-2 variants, including G614, Alpha, Beta, Gamma, Delta, Epsilon, Zeta, Kappa, Lambda, B.1.1.298, B.1.222, B.1.5, and R.1. Both mAbs (especially 17A7) also exhibited strong in vivo efficacy in protecting K18-hACE2 transgenic mice from the lethal infection with G614, Alpha, Beta, Gamma, and Delta viruses. Structural analysis indicated that 17A7 and 17B10 target the tip of the receptor binding motif in the RBD-up conformation. A third RBD-reactive mAb (3A6) although escaped by Beta and Gamma, was highly effective in cross-neutralizing Delta and Omicron BA.1 variants in vitro and in vivo. In competition experiments, antibodies targeting epitopes similar to these 3 mAbs were rarely enriched in human COVID-19 convalescent sera or postvaccination sera. These results are helpful to inform new antibody/vaccine design and these mAbs can be useful tools for characterizing SARS-CoV-2 variants and elicited antibody responses.
Keywords: RBD-reactive antibody; SARS-CoV-2; cross-neutralization; neutralizing antibody; receptor binding domain (RBD); structural analysis.
© 2023 The Authors. Journal of Medical Virology published by Wiley Periodicals LLC.
Conflict of interest statement
CONFLICT OF INTEREST DISCLOUSRE
The authors declare no competing interests.
Figures
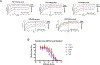


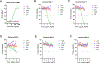



References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

