TIP60 is required for tumorigenesis in non-small cell lung cancer
- PMID: 36916958
- PMCID: PMC10236639
- DOI: 10.1111/cas.15785
TIP60 is required for tumorigenesis in non-small cell lung cancer
Abstract
Histone modifications play crucial roles in transcriptional activation, and aberrant epigenetic changes are associated with oncogenesis. Lysine (K) acetyltransferases 5 (TIP60, also known as KAT5) is reportedly implicated in cancer development and maintenance, although its function in lung cancer remains controversial. Here we demonstrate that TIP60 knockdown in non-small cell lung cancer cell lines decreased tumor cell growth, migration, and invasion. Furthermore, analysis of a mouse lung cancer model with lung-specific conditional Tip60 knockout revealed suppressed tumor formation relative to controls, but no apparent effects on normal lung homeostasis. RNA-seq and ChIP-seq analyses of inducible TIP60 knockdown H1975 cells relative to controls revealed transglutaminase enzyme (TGM5) as downstream of TIP60. Investigation of a connectivity map database identified several candidate compounds that decrease TIP60 mRNA, one that suppressed tumor growth in cell culture and in vivo. In addition, TH1834, a TIP60 acetyltransferase inhibitor, showed comparable antitumor effects in cell culture and in vivo. Taken together, suppression of TIP60 activity shows tumor-specific efficacy against lung cancer, with no overt effect on normal tissues. Our work suggests that targeting TIP60 could be a promising approach to treating lung cancer.
Keywords: KAT5; TGM5; TIP60; artemisinin; lung cancer.
© 2023 The Authors. Cancer Science published by John Wiley & Sons Australia, Ltd on behalf of Japanese Cancer Association.
Conflict of interest statement
SSK reports research support from Boehringer Ingelheim, Johnson & Johnson, MiRXES, and Taiho Therapeutics, as well as personal fees (honoraria) from Boehringer Ingelheim, Bristol Meyers Squibb, and Takeda Pharmaceuticals, all outside the submitted work. HW reports a personal fee (honorarium) from AstraZeneca outside the submitted work. DBC reports receiving consulting fees and honoraria from Takeda/Millennium Pharmaceuticals, AstraZeneca, Pfizer, Blueprint Medicines, and Janssen, institutional research support from Takeda/Millennium Pharmaceuticals, AstraZeneca, Pfizer, Merck Sharp and Dohme, Merrimack Pharmaceuticals, Bristol Myers Squibb, Clovis Oncology, Spectrum Pharmaceuticals, Tesaro and Daiichi Sankyo, and consulting fees from Teladoc and Grand Rounds by Included Health, all outside the submitted work. No other conflict of interest is reported.
Figures
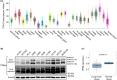

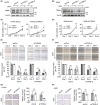
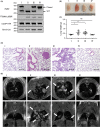

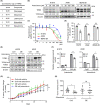

References
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7‐33. - PubMed
-
- Verdone L, Agricola E, Caserta M, Di Mauro E. Histone acetylation in gene regulation. Brief Funct Genomic Proteomic. 2006;5:209‐221. - PubMed
-
- Sapountzi V, Logan IR, Robson CN. Cellular functions of TIP60. Int J Biochem Cell Biol. 2006;38:1496‐1509. - PubMed
-
- Squatrito M, Gorrini C, Amati B. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol. 2006;16:433‐442. - PubMed
-
- Tang Y, Luo J, Zhang W, Gu W. Tip60‐dependent acetylation of p53 modulates the decision between cell‐cycle arrest and apoptosis. Mol Cell. 2006;24:827‐839. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

