Tumor PD-L1 engages myeloid PD-1 to suppress type I interferon to impair cytotoxic T lymphocyte recruitment
- PMID: 36917954
- PMCID: PMC10150625
- DOI: 10.1016/j.ccell.2023.02.005
Tumor PD-L1 engages myeloid PD-1 to suppress type I interferon to impair cytotoxic T lymphocyte recruitment
Abstract
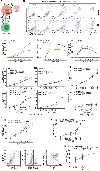
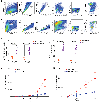
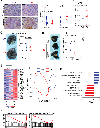
The cellular and molecular mechanisms underlying tumor cell PD-L1 (tPD-L1) function in tumor immune evasion are incompletely understood. We report here that tPD-L1 does not suppress cytotoxic T lymphocyte (CTL) activity in co-cultures of tumor cells and tumor-specific CTLs and exhibits no effect on primary tumor growth. However, deleting tPD-L1 decreases lung metastasis in a CTL-dependent manner in tumor-bearing mice. Depletion of myeloid cells or knocking out PD-1 in myeloid cells (mPD-1) impairs tPD-L1 promotion of tumor lung metastasis in mice. Single-cell RNA sequencing (scRNA-seq) reveals that tPD-L1 engages mPD-1 to activate SHP2 to antagonize the type I interferon (IFN-I) and STAT1 pathway to repress Cxcl9 and impair CTL recruitment to lung metastases. Human cancer patient response to PD-1 blockade immunotherapy correlates with IFN-I response in myeloid cells. Our findings determine that tPD-L1 engages mPD-1 to activate SHP2 to suppress the IFN-I-STAT1-CXCL9 pathway to impair CTL tumor recruitment in lung metastasis.
Keywords: CTL; CXCL9; PD-1; PD-L1; SHP2; immune checkpoint blockade; lung metastasis; type I interferon.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





References
-
- Siddiqui I, Schaeuble K, Chennupati V, Fuertes Marraco SA, Calderon-Copete S, Pais Ferreira D, Carmona SJ, Scarpellino L, Gfeller D, Pradervand S, et al. (2019). Intratumoral Tcf1+PD-1+CD8+ T Cells with Stem-like Properties Promote Tumor Control in Response to Vaccination and Checkpoint Blockade Immunotherapy. Immunity 50, 195–211.e110. 10.1016/j.immuni.2018.12.021. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

