STING signalling is terminated through ESCRT-dependent microautophagy of vesicles originating from recycling endosomes
- PMID: 36918692
- PMCID: PMC10014584
- DOI: 10.1038/s41556-023-01098-9
STING signalling is terminated through ESCRT-dependent microautophagy of vesicles originating from recycling endosomes
Abstract
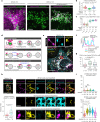
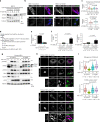
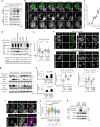
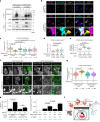
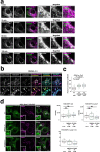
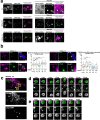
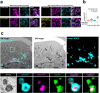
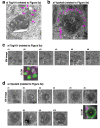
Stimulator of interferon genes (STING) is essential for the type I interferon response against a variety of DNA pathogens. Upon emergence of cytosolic DNA, STING translocates from the endoplasmic reticulum to the Golgi where STING activates the downstream kinase TBK1, then to lysosome through recycling endosomes (REs) for its degradation. Although the molecular machinery of STING activation is extensively studied and defined, the one underlying STING degradation and inactivation has not yet been fully elucidated. Here we show that STING is degraded by the endosomal sorting complexes required for transport (ESCRT)-driven microautophagy. Airyscan super-resolution microscopy and correlative light/electron microscopy suggest that STING-positive vesicles of an RE origin are directly encapsulated into Lamp1-positive compartments. Screening of mammalian Vps genes, the yeast homologues of which regulate Golgi-to-vacuole transport, shows that ESCRT proteins are essential for the STING encapsulation into Lamp1-positive compartments. Knockdown of Tsg101 and Vps4, components of ESCRT, results in the accumulation of STING vesicles in the cytosol, leading to the sustained type I interferon response. Knockdown of Tsg101 in human primary T cells leads to an increase the expression of interferon-stimulated genes. STING undergoes K63-linked ubiquitination at lysine 288 during its transit through the Golgi/REs, and this ubiquitination is required for STING degradation. Our results reveal a molecular mechanism that prevents hyperactivation of innate immune signalling, which operates at REs.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures










Comment in
-
STING is ESCRTed to degradation by microautophagy.Nat Cell Biol. 2023 Mar;25(3):379-380. doi: 10.1038/s41556-022-01084-7. Nat Cell Biol. 2023. PMID: 36918691 No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

