A multi-organoid platform identifies CIART as a key factor for SARS-CoV-2 infection
- PMID: 36918693
- PMCID: PMC10014579
- DOI: 10.1038/s41556-023-01095-y
A multi-organoid platform identifies CIART as a key factor for SARS-CoV-2 infection
Abstract
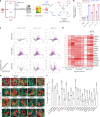
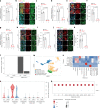
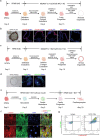
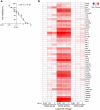
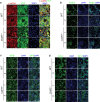
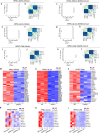
COVID-19 is a systemic disease involving multiple organs. We previously established a platform to derive organoids and cells from human pluripotent stem cells to model SARS-CoV-2 infection and perform drug screens1,2. This provided insight into cellular tropism and the host response, yet the molecular mechanisms regulating SARS-CoV-2 infection remain poorly defined. Here we systematically examined changes in transcript profiles caused by SARS-CoV-2 infection at different multiplicities of infection for lung airway organoids, lung alveolar organoids and cardiomyocytes, and identified several genes that are generally implicated in controlling SARS-CoV-2 infection, including CIART, the circadian-associated repressor of transcription. Lung airway organoids, lung alveolar organoids and cardiomyocytes derived from isogenic CIART-/- human pluripotent stem cells were significantly resistant to SARS-CoV-2 infection, independently of viral entry. Single-cell RNA-sequencing analysis further validated the decreased levels of SARS-CoV-2 infection in ciliated-like cells of lung airway organoids. CUT&RUN, ATAC-seq and RNA-sequencing analyses showed that CIART controls SARS-CoV-2 infection at least in part through the regulation of NR4A1, a gene also identified from the multi-organoid analysis. Finally, transcriptional profiling and pharmacological inhibition led to the discovery that the Retinoid X Receptor pathway regulates SARS-CoV-2 infection downstream of CIART and NR4A1. The multi-organoid platform identified the role of circadian-clock regulation in SARS-CoV-2 infection, which provides potential therapeutic targets for protection against COVID-19 across organ systems.
© 2023. The Author(s).
Conflict of interest statement
R.E.S. is on the scientific advisory board of Miromatrix Inc, and is a paid consultant and speaker for Alnylam Inc. S.C. and T.E. are the co-founders of OncoBeat, LLC. S.C. is a consultant for Vesalius Therapeutics. The remaining authors declare no competing interests.
Figures






Comment in
-
Linking SARS-CoV-2 to the circadian clock.Nat Cell Biol. 2023 Mar;25(3):373-374. doi: 10.1038/s41556-023-01099-8. Nat Cell Biol. 2023. PMID: 36918694 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

