Cancer-associated fibroblast-derived secreted phosphoprotein 1 contributes to resistance of hepatocellular carcinoma to sorafenib and lenvatinib
- PMID: 36919193
- PMCID: PMC10091107
- DOI: 10.1002/cac2.12414
Cancer-associated fibroblast-derived secreted phosphoprotein 1 contributes to resistance of hepatocellular carcinoma to sorafenib and lenvatinib
Abstract
Background: Cancer-associated fibroblasts (CAFs) play an important role in the induction of chemo-resistance. This study aimed to clarify the mechanism underlying CAF-mediated resistance to two tyrosine kinase inhibitors (TKIs), sorafenib and lenvatinib, and to identify a novel therapeutic target for overcoming TKI resistance in hepatocellular carcinoma (HCC).
Methods: We performed a systematic integrative analysis of publicly available gene expression datasets and whole-transcriptome sequencing data from 9 pairs of CAFs and para-cancer fibroblasts isolated from human HCC and para-tumor tissues, respectively, to identify key molecules that might induce resistance to TKIs. We then performed in vitro and in vivo experiments to validate selected targets and related mechanisms. The associations of plasma secreted phosphoprotein 1 (SPP1) expression levels before sorafenib/lenvatinib treatment with progression-free survival (PFS) and overall survival (OS) of 54 patients with advanced HCC were evaluated using Kaplan-Meier and Cox regression analysis.
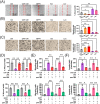
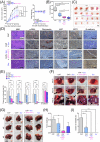
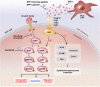
Results: Bioinformatic analysis identified CAF-derived SPP1 as a candidate molecule driving TKI resistance. SPP1 inhibitors reversed CAF-induced TKI resistance in vitro and in vivo. CAF-derived SPP1 activated rapidly accelerated fibrosarcoma (RAF)/mitogen-activated protein kinase (MAPK) and phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT)/mammalian target of rapamycin (mTOR) through the integrin-protein kinase C-alpha (PKCα) signaling pathway and promoted epithelial-to-mesenchymal transition (EMT). A high plasma SPP1 level before TKI treatment was identified as an independent predictor of poor PFS (P = 0.026) and OS (P = 0.047) in patients with advanced HCC after TKI treatment.
Conclusions: CAF-derived SPP1 enhances TKI resistance in HCC via bypass activation of oncogenic signals and EMT promotion. Its inhibition represents a promising therapeutic strategy against TKI resistance in HCC. Moreover, plasma SPP1 level before TKI treatment represents a potential biomarker for treatment response prediction.
Keywords: drug resistance; epithelial-to-mesenchymal transition; hepatocellular carcinoma; secreted phosphoprotein 1.
© 2023 The Authors. Cancer Communications published by John Wiley & Sons Australia, Ltd. on behalf of Sun Yat-sen University Cancer Center.
Conflict of interest statement
The authors have declared that no conflict of interest exists.
Figures






References
-
- European Association for the Study of the Liver. Electronic address eee, European Association for the Study of the L. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J Hepatol. 2018;69(1):182‐236. - PubMed
-
- Llovet JM, Villanueva A, Marrero JA, Schwartz M, Meyer T, Galle PR, et al. Trial Design and Endpoints in Hepatocellular Carcinoma: AASLD Consensus Conference. Hepatology. 2021;73 Suppl 1:158‐91. - PubMed
-
- Cheng AL, Hsu C, SL Chan, Choo SP, Kudo M. Challenges of combination therapy with immune checkpoint inhibitors for hepatocellular carcinoma. J Hepatol. 2020;72(2):307‐19. - PubMed
-
- Faivre S, Rimassa L, Finn RS. Molecular therapies for HCC: Looking outside the box. J Hepatol. 2020;72(2):342‐52. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

