F-actin-rich territories coordinate apoptosome assembly and caspase activation during DNA damage-induced intrinsic apoptosis
- PMID: 36920061
- PMCID: PMC10162420
- DOI: 10.1091/mbc.E22-04-0119
F-actin-rich territories coordinate apoptosome assembly and caspase activation during DNA damage-induced intrinsic apoptosis
Abstract
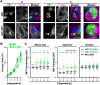
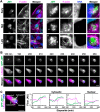
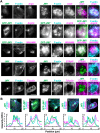
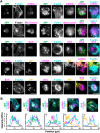
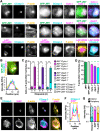
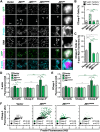
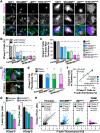
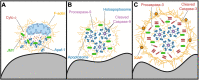
The actin cytoskeleton is a ubiquitous participant in cellular functions that maintain viability, but how it controls programmed cell death is not well understood. Here we show that in response to DNA damage, human cells form a juxtanuclear F-actin-rich territory that coordinates the organized progression of apoptosome assembly to caspase activation. This cytoskeletal compartment is created by the actin nucleation factors JMY, WHAMM, and the Arp2/3 complex, and it excludes proteins that inhibit JMY and WHAMM activity. Within the territory, mitochondria undergo outer membrane permeabilization and JMY localization overlaps with punctate structures containing the core apoptosome components cytochrome c and Apaf-1. The F-actin-rich area also encompasses initiator caspase-9 and clusters of a cleaved form of executioner caspase-3 but restricts accessibility of the caspase inhibitor XIAP. The clustering and potency of caspase-3 activation are positively regulated by the amount of actin polymerized by JMY and WHAMM. These results indicate that JMY-mediated actin reorganization functions in apoptotic signaling by coupling the biogenesis of apoptosomes to the localized processing of caspases.
Figures

References
-
- Ahn JS, Jang IS, Kim DI, Cho KA, Park YH, Kim K, Kwak CS, Chul Park S (2003). Aging-associated increase of gelsolin for apoptosis resistance. Biochem Biophys Res Commun 312, 1335–1341. - PubMed
-
- Bock FJ, Tait SWG (2020). Mitochondria as multifaceted regulators of cell death. Nat Rev Mol Cell Biol 21, 85–100. - PubMed
-
- Bolte S, Cordelières FP (2006). A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224, 213–232. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

