Centromere proteins are asymmetrically distributed between newly divided germline stem and daughter cells and maintain a balanced niche in Drosophila males
- PMID: 36920070
- PMCID: PMC10162413
- DOI: 10.1091/mbc.E22-10-0466
Centromere proteins are asymmetrically distributed between newly divided germline stem and daughter cells and maintain a balanced niche in Drosophila males
Abstract
Stem cells can undergo asymmetric cell division (ACD) giving rise to one new stem cell and one differentiating daughter cell. In Drosophila germline stem cells (GSCs), the centromeric histone CENP-A (CID in flies) is asymmetrically distributed between sister chromatids such that chromosomes that end up in the GSC harbor more CID at centromeres. A model of "mitotic drive" has been proposed in GSCs such that stronger and earlier centromere and kinetochore interactions with microtubules bias sister chromatid segregation. Here we show that in Drosophila males, centromere proteins CID, CAL1, and CENP-C are asymmetrically distributed in newly divided GSCs and daughter cells in S phase. We find that overexpression of CID (either with or without CAL1) or CENP-C depletion disrupts CID asymmetry, with an increased pool of GSCs relative to daughter cells detectable in the niche. This result suggests a shift toward GSC self-renewal rather than differentiation, important for maintaining tissue homeostasis. Overexpression of CAL1 does not disrupt asymmetry, but instead drives germ cell proliferation in the niche. Our results in male GSCs are comparable to female GSCs, indicating that despite differences in signaling, organization, and niche composition, the effects of centromere proteins on GSC maintenance are conserved between the sexes.
Figures

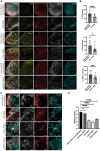




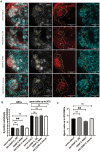
References
-
- Bade D, Pauleau AL, Wendler A, Erhardt S (2014). The E3 ligase CUL3/RDX controls centromere maintenance by ubiquitylating and stabilizing CENP-A in a CAL1-dependent manner. Dev Cell 28, 508–519. - PubMed
-
- Carty BL, Dunleavy EM (2020). Centromere assembly and non-random sister chromatid segregation in stem cells. Essays Biochem 64, 223–232. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

