Distinct Aurora B pools at the inner centromere and kinetochore have different contributions to meiotic and mitotic chromosome segregation
- PMID: 36920098
- PMCID: PMC10162408
- DOI: 10.1091/mbc.E23-01-0014
Distinct Aurora B pools at the inner centromere and kinetochore have different contributions to meiotic and mitotic chromosome segregation
Abstract
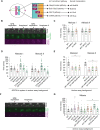
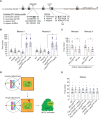
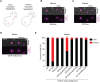
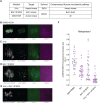
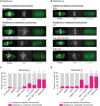
Proper chromosome segregation depends on the establishment of bioriented kinetochore-microtubule attachments, which often requires multiple rounds of release and reattachment. Aurora B and C kinases phosphorylate kinetochore proteins to release tensionless attachments. Multiple pathways recruit Aurora B/C to the centromere and kinetochore. We studied how these pathways contribute to anaphase onset timing and correction of kinetochore-microtubule attachments in budding yeast meiosis and mitosis. We find that the pool localized by the Bub1/Bub3 pathway sets the normal duration of meiosis and mitosis, in differing ways. Our meiosis data suggests a model that disruption of this pathway leads to PP1 kinetochore localization, which dephosphorylates Cdc20 for premature anaphase onset. For error correction, the Bub1/Bub3 and COMA pathways are individually important in meiosis but compensatory in mitosis. Finally, we find that the haspin and Bub1/3 pathways function together to ensure error correction in mouse oogenesis. Our results suggest that each recruitment pathway localizes spatially distinct kinetochore-localized Aurora B/C pools that function differently between meiosis and mitosis.
Figures

Update of
-
Distinct Aurora B pools at the inner centromere and kinetochore have different contributions to meiotic and mitotic chromosome segregation.bioRxiv [Preprint]. 2023 Feb 5:2023.02.05.527197. doi: 10.1101/2023.02.05.527197. bioRxiv. 2023. Update in: Mol Biol Cell. 2023 May 1;34(5):ar43. doi: 10.1091/mbc.E23-01-0014. PMID: 36778459 Free PMC article. Updated. Preprint.
References
-
- Avo Santos M, van de Werken C, de Vries M, Jahr H, Vromans MJM, Laven JSE, Fauser BC, Kops GJ, Lens SM, Baart EB. (2011). A role for Aurora C in the chromosomal passenger complex during human preimplantation embryo development. Hum Reprod 26, 1868–81. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

